Transposable elements can be described as mobile genetic segments that is able to move from one position of the genome to another without the need of any homology. It is the process where small DNA sequences is shifted or sometimes replicated inside the chromosomes, and this movement is referred to as transposition.
It is seen that these elements are present in almost all living organisms and a major part of the eukaryotic genome is formed by such repetitive and mobile DNA. In some plants these elements occupy a very large percentage of the total DNA. It is the process where the movement of the element may produce different chromosomal changes like deletions, inversions or sometimes fusion of chromosomes, and in this way genetic variations is introduced.
Two major groups of transposable elements is Class I or retrotransposons which move by a copy and paste mechanism and Class II or DNA transposons which move by a cut and paste mechanism where the enzyme transposase is involved. These are also responsible for carrying antimicrobial resistance genes in bacteria, and the element may insert randomly into plasmids or chromosomes. It can also move between bacteria by conjugation, transformation or transduction.
The major source of the discovery of transposable elements was the study of genetic instability in maize by Barbara McClintock (1965).
Definition of Transposable elements
Transposable elements are small mobile DNA sequences that is able to move from one genomic location to another, and this movement is referred to as transposition in the genome.
Characteristics of transposable elements
Transposable elements, also known as transposons, possess several characteristic features:
- Transposable elements are segments of DNA that can shift their position within the genome and it is commonly referred to as “jumping genes”.
- It is present in almost all prokaryotic as well as eukaryotic genomes and it occupies a major portion of the genome in many organisms.
- These elements is grouped into two major classes– Class I (Retrotransposons) and Class II (DNA transposons). Class I uses RNA intermediate while Class II uses DNA intermediate.
- Class I elements move by a copy–paste method. It is transcribed into RNA and again converted to DNA (by reverse transcriptase) which is inserted at a new location. The original copy remains in the genome.
- Retrotransposons is of two types– LTR elements and non-LTR elements (LINEs and SINEs). These are common in higher organisms and LINE-1 is one active autonomous element in humans.
- SINEs (like Alu) are non-autonomous elements which depend on LINE enzymes for their movement.
- Class II elements move by a cut–paste method. The transposase enzyme excises the element and inserts it into another genomic site.
- These DNA transposons have terminal inverted repeats (TIRs) which are recognised by transposase. The excised DNA is then integrated into another site.
- Some special DNA transposons like Helitrons move by a rolling circle mechanism while Mavericks/Polintons are self-synthesizing DNA elements.
- During insertion of transposable elements, short sequences called target site duplications (TSDs) is produced. These are marks used to identify past transposition events.
- Transposable elements may be autonomous (having their own enzyme activity like transposase or reverse transcriptase) or non-autonomous (without enzymatic activity). These use host or other TE enzymes.
- It is known that transposable elements cause mutation by inserting into coding or regulatory region, causing disruption or altered expression of genes.
- These elements also induce chromosomal rearrangements because recombination can occur between repeated TE sequences.
- To suppress harmful activity, organisms use DNA methylation and small RNA mechanisms (like piRNA) which silence TE movement.
- Some TEs is utilised beneficially during evolution. For example, RAG genes for V(D)J recombination in vertebrates are derived from ancient DNA transposons.
- Among the important modern discoveries, CRISPR-associated transposons use CRISPR-Cas components for targeted insertion of DNA at specific genomic sites.
Types of Transposable elements
Transposable elements are of the following two types:
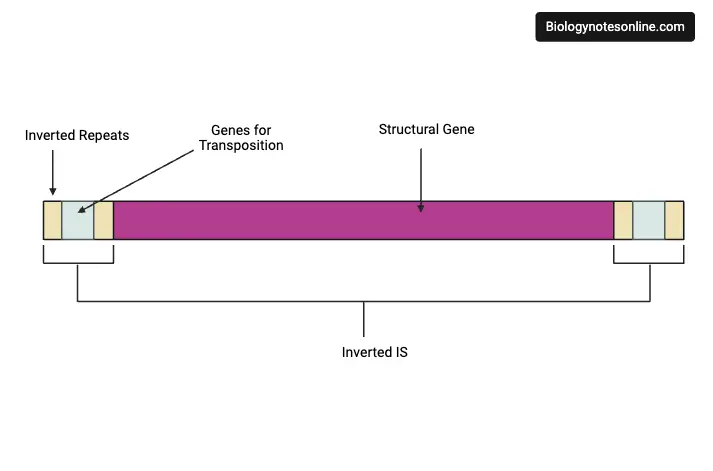
1. Insertion Sequences (IS) or Simple Transposons
- It is the simplest type of transposable element.
- These sequences are short (800–1500 bp) and it does not code for proteins.
- It carries only the genetic information required for its transposition, mainly the gene for transposase.
- It is identified in bacteriophages, in F factor plasmid and many bacteria.
- These are usually flanked by short inverted repeat sequences which help in the transposition process.
2. Transposons (Tn) or Complex Transposons
- These are several thousand base pair long and it contains genes that code for one or more proteins, including antibiotic resistance factors in bacteria.
- A distinguishing feature is the presence of identical inverted terminal repeats (IR) ranging from 8 to 38 bp.
- These IR sequences are characteristic for different types of transposons.
- A short sequence (less than 10 bp) is present on both sides of the transposon which becomes duplicated during insertion.
- The insertion of transposon causes duplication of a single target sequence, appearing as direct repeats flanking the inserted element.
- The flanking direct repeats are not considered as part of the transposon and these segments act like IS or IS-like regions.
Mechanism of Transposition
It is the principle where a limited number of nuclease domains catalyze the cleavage and joining of DNA. The domains are RNase H-like (DDE), HUH nuclease domains, serine recombinase and tyrosine recombinase domains. It is these catalytic motifs that decide the chemical mechanism used in transposition.
Types of Transposition Mechanisms
Some of the main types are–
- RNase H-like (DDE) transposition
- HUH transposition
- Retrotransposon mediated transposition
- CRISPR-associated transposition
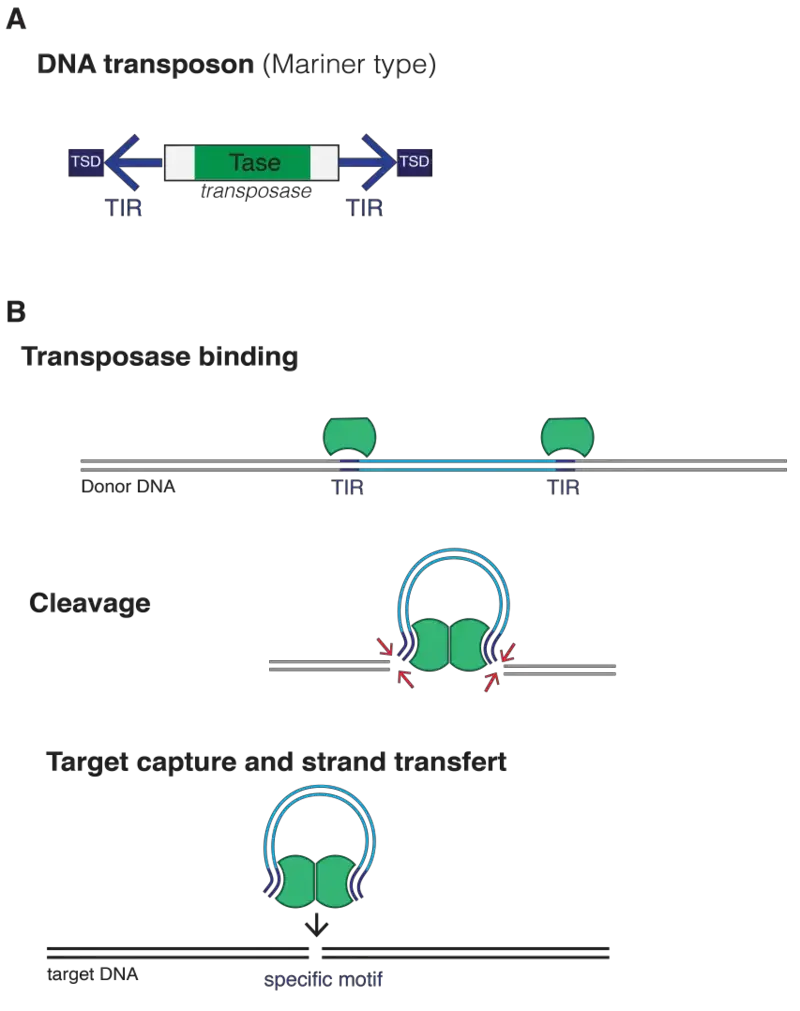
1. RNase H-like (DDE) Transposases
It is the most common catalytic domain present in transposases of Tc1/mariner, hAT, piggyBac superfamilies and also retroviral integrases.
Catalytic Core
It is composed of conserved acidic residues forming the DDE or DDD motif. These residues bind two divalent metal ions (Mg²⁺ or Mn²⁺). These ions activate water molecules or 3′-OH groups, and nucleophilic attack occurs on the phosphodiester bond.
Synaptic Complex Formation
In this step the transposase binds to the inverted terminal repeats (ITRs). These ends is brought together and form a synaptic complex (transpososome). The reaction is controlled so that cleavage takes place only when both ends is paired. In many cases the enzyme act in trans, where one subunit bound to one end cleaves the other end.
Excision (Cut-and-Paste)
The reaction is as follows–
First the enzyme hydrolyses the phosphodiester bonds and form free 3’-OH groups.
Two strategies are commonly observed–
- Direct cleavage – both strands is hydrolysed sequentially (Tc1/mariner).
- Hairpin formation – the 3’-OH attack form a hairpin which is later resolved (Tn5 and piggyBac).
Integration
The excised element inserts into target DNA by transesterification. The 3’-OH ends attack the target at staggered positions. After this host repair system fills the gaps forming Target Site Duplications (TSDs).
2. HUH Transposases (Single-Stranded Transposition)
It is the mechanism used by Helitrons and IS200/IS605 family. The domain has two histidines and one hydrophobic residue.
Mechanism
A tyrosine residue attack the phosphodiester backbone and forms a 5′-phosphotyrosine intermediate.
Rolling Circle / Peel-and-Paste
Helitrons move by a rolling circle mechanism where the leading strand is peeled and copied. IS200 family produce excised ssDNA circles. No TSDs is formed in this case.
3. Retrotransposon Mechanisms (Class I)
Retrotransposons move by an RNA intermediate. It is the copy-and-paste mechanism.
LTR Retrotransposons
The DNA is transcribed into RNA which is reverse-transcribed to cDNA. Integrase (similar to DDE transposases) inserts the cDNA.
Non-LTR Retrotransposons (TPRT)
LINE-1 and Alu use Target-Primed Reverse Transcription. The endonuclease makes a nick exposing 3′-OH. This OH works as primer for reverse transcription of retrotransposon RNA at the target site.
4. CRISPR-Associated Transposition (CAST)
It is controlled by Tn7-like systems. These systems combine CRISPR targeting with transposition.
Recruitment
CRISPR effector (Cascade-like complex) bind the target DNA using guide RNA. It is the process which replace the normal DNA-binding by transposase.
Activation
After recognition, TniQ is recruited. It then recruit TnsC which polymerises on DNA. This complex brings TnsA and TnsB, and insertion is done at a defined region downstream of the target site.
Among the important consequences of transposition is genome rearrangement, gene duplication and generation of mutations. It also contributes to horizontal gene transfer and genome evolution.
Methods for Transposable elements (TE) detection
1. Computational and Bioinformatic Detection
This is the most used method for scanning genome sequences. It is based on analysing repeated DNA sequences and classifying them.
De novo Repeat Identification
It is the process where repeats is searched without any reference. The main steps are–
- Finding the repeated regions
- Preparing the consensus sequence
- Classifying the repeat family
Algorithms Used
Some of the important algorithms are–
- K-mer Method
It scans the genome for short sequences (k length) which occur more than expected. Mismatches is allowed and extension is done until similarity ends. - Sequence Self-Comparison
The genome sequence is aligned with itself. It helps in detecting diverged or fragmented transposons. - Periodicity Method
Fourier transformation is applied to detect periodic patterns. Peaks in the spectrum indicate repeated elements. It is slow for whole genome level.
Consensus Building
All copies of a repeat family is aligned and the most common base at each position is taken. This sequence represent the ancestral TE sequence.
Databases and Tools
Some of the important tools are–
- RepeatMasker
- RepeatModeler2
- Repbase
- Dfam
- RepetDB
These are used for annotation and classification of TEs.
2. Sequencing-Based Technologies (Tn5 System)
It is the method where a hyperactive Tn5 transposase is used to cut DNA and insert adapters. This reaction is referred to as tagmentation.
ATAC-seq
It is the process used to study open chromatin. Tn5 inserts into DNA which is accessible. It help in studying chromatin accessibility, nucleosome position and protein binding.
Some variants are–
- scATAC-seq (single cell)
- ATAC-me (with methylation detection)
Visualization Techniques
Modified Tn5 with fluorescent adapters is used in ATAC-see and 3D ATAC-PALM. It allow direct viewing of open chromatin under microscope.
LIANTI Method
Linear amplification using Tn5 where T7 promoter is inserted. It is used for single-cell genome sequencing and detection of single nucleotide variation.
3. Experimental and Genetic Screening Methods
Engineered transposons is used in living organisms for functional studies and mutational screening.
Insertional Mutagenesis
Transposons like Sleeping Beauty (SB) and piggyBac (PB) is introduced to create mutations. Insertion sites in tumors or mutants is mapped to detect important genes.
Gene Traps
It is the process where a splice acceptor and reporter gene is placed inside a TE. When inserted inside a gene, the reporter is expressed. It detect the expression pattern of disrupted genes.
Enhancer Traps
A minimal promoter and reporter gene is inserted inside the transposon. Insertion near an enhancer activates the reporter and show the tissue-specific activity.
Mapping insertion sites
PCR-based methods like splinkerette PCR is used to clone flanking genomic sequences and identify the targeted genes.
4. Structural Visualization
This is the method used to study TE complexes at molecular level.
Cryo-Electron Microscopy (Cryo-EM)
It is used to determine structures of transposition complexes like Cascade–TniQ–TnsC in CRISPR-associated transposons. It shows how the TE machinery recognises DNA and recruits the transposase.
Examples of Transposable elements
Class I Retrotransposons (RNA-mediated)
Some of the important examples are–
- LINEs (Long Interspersed Nuclear Elements)
– LINE-1 (L1)
– R2 elements - SINEs (Short Interspersed Nuclear Elements)
– Alu elements
– B1 elements - LTR Retrotransposons / Endogenous Retroviruses
– Ty elements (Ty1, Ty3, Ty5)
– Copia
– IAP (Intracisternal A-particle)
Class II DNA Transposons (Cut-and-Paste)
Some of the common examples are–
- Ac/Ds System (Maize)
– Activator (Ac)
– Dissociation (Ds) - P Elements (Drosophila)
- Tc1/Mariner Superfamily
– Sleeping Beauty (SB)
– Tc1
– Mos1
– Minos - piggyBac Transposon
- hAT Superfamily
– Tol2
– Hermes - Bacterial Transposons
– Tn5
– Tn7
– Tn10
– Mu (Bacteriophage Mu) - Helitrons (Rolling-circle elements)
CRISPR-Associated Transposons (CASTs)
Some of the important examples are–
- Tn6677 (VchCAST)
- Tn7747 (ShCAST)
Diseases Caused by Transposable Elements (TEs)
Genetic and Hereditary Disorders
Some of the important diseases caused by germline TE insertions are–
- Hemophilia A– -Caused by LINE-1 insertion inside the Factor VIII gene.
- Hemophilia B– Insertions of Alu, SVA or LINE-1 inside the Factor IX gene.
- Duchenne Muscular Dystrophy– TE insertion disrupts the dystrophin gene.
- Fukuyama Congenital Muscular Dystrophy (FCMD)-SVA retrotransposon insertion in the FKTN gene.
- Neurofibromatosis Type 1– New Alu insertions inside the NF1 gene.
- Retinitis Pigmentosa– Homozygous Alu insertion in the MAK gene affecting splicing.
- Lesch–Nyhan Syndrome– Alu-mediated recombination causing deletion or duplication in HPRT gene.
- Apert Syndrome– TE-associated mutations affecting FGFR2 gene.
- Familial Hypercholesterolemia– TE insertion causing gene disruption in LDL receptor region.
- Metabolic and Immune Disorders– TE involvement is linked with porphyria, SCID and chronic granulomatous disease.
Cancer Associated with TE Insertions
Transposable elements is activated in many cancers through somatic insertions. Some of the common associations are–
- Colorectal Cancer-LINE-1 insertion in APC gene causing disruption.
- Breast Cancer– Alu and L1 insertions affecting BRCA1 gene expression.
- Ovarian Cancer– Alu-mediated exon loss from BRCA2 gene.
- Esophageal Adenocarcinoma– High somatic LINE-1 retrotransposition observed.
- Head and Neck Carcinoma– Increased TE activity causing instability.
- Lung Squamous Carcinoma– TE insertions contributing to genomic alterations.
- Epithelial Tumors– LINE-1 activity linked with poor survival in APC-related cases.
Autoimmune and Inflammatory Diseases
Some diseases arise when TE RNA or DNA accumulate and activate immune pathways.
- Aicardi–Goutières Syndrome (AGS)– Mutations in genes that suppress TEs (TREX1, SAMHD1, ADAR1). TE-derived nucleic acids activate the cGAS–STING pathway.
- Inflammaging (Age-related Inflammation)– Loss of heterochromatin causes activation of TEs with age.
- Neurodegeneration– TE activation is linked with Alzheimer’s disease and other tauopathies.
Genome Instability Disorders Caused by TEs
Some of the effects caused by TE-driven structural alterations are–
- Non-Allelic Homologous Recombination (NAHR)– Alu-mediated misalignment causing deletions or duplications.
- Double-Strand Breaks (DSBs)– L1 endonuclease activity causes DNA cleavage leading to instability.
Negative Effects of Transposable Elements (TEs)
Insertional Mutagenesis and Gene Disruption
Some of the main negative effects are–
- Disruption of Coding Sequence– When a TE inserts inside an exon, the gene function is lost. It may change the reading frame or introduce a stop codon.
- Interference in Splicing– Insertions inside introns alter the normal splice sites. It can produce improper splicing or exon skipping (e.g., Alu insertion affecting BRCA2).
- Regulatory Region Disturbance– TEs bring their own promoters or enhancers. Insertion near 5’ or 3’ regions alter gene expression and stability.
Genomic Instability and Structural Rearrangements
These elements cause instability by their repetitive and mobile nature.
- Non-Allelic Homologous Recombination (NAHR)– Similar TE copies misalign. It result in large deletions, duplications, inversions or translocations.
- Double-Strand Break Formation– Cut-and-paste elements leave gaps which must be repaired. LINE-1 endonuclease also produces frequent double-strand breaks.
- Hairpin Structure Formation– Inverted repeats form secondary structures. These regions become breakage hotspots causing chromosome instability.
Association with Human Diseases
Transposable element activity is linked with many disorders.
- Genetic Disorders
– Hemophilia A and B
– Duchenne muscular dystrophy
– Porphyria
– Neurofibromatosis
– X-linked SCID - Cancer Development
– Insertion inside tumor suppressor genes like APC and PTEN.
– LINE-1 activation in colorectal and esophageal cancers.
– Hypomethylation reactivates TE promoters causing abnormal transcripts. - Neurological Disorders
– TE activation is linked with Alzheimer’s disease and other tauopathies.
Inflammatory and Aging Effects
These elements produce sterile inflammation when not properly silenced.
- Induction of Sterile Inflammation– TE-derived DNA or RNA accumulates in cytoplasm. It activate cGAS–STING pathway similar to viral infection.
- Autoimmune Effects– Persistent TE activation produce type I interferon response. Aicardi–Goutières syndrome is commonly associated.
- Age-related Effects– Loss of heterochromatin in aging cells activate TEs. It contribute to inflammaging and genomic instability.
Metabolic Cost and Selfish DNA Behaviour
Some harmful effects arise from the resource usage of TEs.
- Resource Consumption– TE transcription and replication use nucleotides and energy.
- Overproduction Inhibition– Excess transposase levels interfere with TE activity. It indicate that too much transposition is harmful for the host cell.
Applications of transposable elements
- Insertional mutagenesis
- It is the process in which the transposable element insert into a gene and disturb its activity.
- These insertions is used to identify unknown genes because the phenotype of the organism is changed after insertion.
- In cancer studies, Sleeping Beauty (SB) and piggyBac (PB) systems are used for detecting oncogenes and tumor suppressor genes.
- Gene trap and enhancer trap elements are designed for studying gene expression because the reporter gene is activated when insertion occur near regulatory regions.
- Genome-wide screening
- Transposable elements is used to create large mutant libraries.
- PiggyBac system is applied in mice and Plasmodium falciparum to find essential genes.
- This is referred to as random mutagenesis screening.
- Transgenesis
- It is the process in which foreign DNA is inserted into the host genome.
- SB, PB and Tol2 elements are used in vertebrate models because these elements can integrate stably into chromosomes.
- These are helpful in producing transgenic lines for developmental studies.
- Gene therapy applications
- The transposable element system act as non-viral vectors for transferring therapeutic genes.
- SB is used for engineering CAR-T cells.
- PB helps in stem cell engineering because the PB element can be excised without leaving any footprint.
- The integration preference of different TE systems is an important feature because SB mostly insert into intergenic regions which is considered safer.
- Genomic sequencing technologies
- Tn5 transposase is used in ATAC-seq. It is the process in which open chromatin regions are tagged by transposase and sequencing adapters are added at the same time.
- Tn5 is also used in LIANTI for single-cell genome amplification.
- Precision genome engineering
- CRISPR-associated transposons (CASTs) are Tn7-like systems which use guide RNA to select the insertion site.
- It is the process where large DNA fragments can be inserted without creating double strand breaks.
- The reaction is carried out by Cas protein complex with Tns proteins.
- Evolutionary role of transposable elements
- Some of the important biological systems in nature evolved from ancient transposons.
- RAG1 and RAG2 proteins, required for V(D)J recombination, originated from Transib transposons.
- CENP-B protein is derived from a pogo-like transposon.
FAQ
- Ayarpadikannan, S., & Kim, H.-S. (2014). The impact of transposable elements in genome evolution and genetic instability and their implications in various diseases. Genomics & Informatics, 12(3), 98–104. https://doi.org/10.5808/GI.2014.12.3.98
- Craig, N. L. (2002). Transposases and integrases. In Encyclopedia of Life Sciences. Nature Publishing Group.
- Han, M., Perkins, M. H., Novaes, L. S., Xu, T., & Chang, H. (2023). Advances in transposable elements: From mechanisms to applications in mammalian genomics. Frontiers in Genetics, 14, 1290146. https://doi.org/10.3389/fgene.2023.1290146
- Hickman, A. B., & Dyda, F. (2015). Mechanisms of DNA transposition. Microbiology Spectrum, 3(2), MDNA3-0034-2014. https://doi.org/10.1128/microbiolspec.MDNA3-0034-2014
- Hsieh, S.-C., & Peters, J. E. (2024). Natural and engineered guide RNA–directed transposition with CRISPR-associated Tn7-like transposons. Annual Review of Biochemistry, 93(1), 139–161. https://doi.org/10.1146/annurev-biochem-030122-041908
- Kimball, J. W. (2025, March 17). 10.4: Transposons – “jumping genes”. Biology LibreTexts. https://bio.libretexts.org/Bookshelves/Introductory_and_General_Biology/Biology_(Kimball)/10%3A_Mutation/10.04%3A_Transposons_-_jumping_genes
- Muñoz-López, M., & García-Pérez, J. L. (2010). DNA transposons: Nature and applications in genomics. Current Genomics, 11(2), 115–128. https://doi.org/10.2174/138920210790886871
- Nesmelova, I. V., & Hackett, P. B. (2010). DDE transposases: Structural similarity and diversity. Advanced Drug Delivery Reviews, 62(12), 1187–1195. https://doi.org/10.1016/j.addr.2010.06.006
- Schmitz, M., & Querques, I. (2024). DNA on the move: Mechanisms, functions and applications of transposable elements. FEBS Open Bio, 14(1), 13–22. https://doi.org/10.1002/2211-5463.13743
- Short interspersed nuclear element. (2025, September 14). In Wikipedia. https://en.wikipedia.org/w/index.php?title=Short_interspersed_nuclear_element&oldid=1314971421
- Transposable element. (2025, September 20). In Wikipedia. https://en.wikipedia.org/w/index.php?title=Transposable_element&oldid=1326540075
- Transposable elements: Drivers of genomic dynamics and precision engineering tools. (n.d.). [Provided source material].
- Wang, S. (2025). Molecular mechanism of the type I-B2 CRISPR-associated transposon system [Doctoral dissertation, Purdue University]. Purdue e-Pubs. https://doi.org/10.25394/PGS.28862345
- Wang, S., Gabel, C., Siddique, R., Klose, T., & Chang, L. (2023). Molecular mechanism for Tn7-like transposon recruitment by a type I-B CRISPR effector. Cell, 186(19), 4204–4215.e19. https://doi.org/10.1016/j.cell.2023.07.010
- Wells, J. N., & Feschotte, C. (2020). A field guide to eukaryotic transposable elements. Annual Review of Genetics, 54, 539–561. https://doi.org/10.1146/annurev-genet-040620-022145
- McClintock, B. (1950). The origin and behavior of mutable loci in maize. Proceedings of the National Academy of Sciences, 36(6), 344-355.
- Feschotte, C. (2008). Transposable elements and the evolution of regulatory networks. Nature Reviews Genetics, 9(5), 397-405.
- Kazazian, H. H. (2004). Mobile elements: drivers of genome evolution. Science, 303(5664), 1626-1632.
- Lander, E. S., Linton, L. M., Birren, B., Nusbaum, C., Zody, M. C., Baldwin, J., … & Task Force on Sequencing the Human Genome. (2001). Initial sequencing and analysis of the human genome. Nature, 409(6822), 860-921.
- Arkhipova, I. R. (2017). Transposable elements in sexual reproduction. Advances in Experimental Medicine and Biology, 1046, 471-491.
- Goodier, J. L., & Kazazian, H. H. (2008). Retrotransposons revisited: the restraint and rehabilitation of parasites. Cell, 135(1), 23-35.
- Bourque, G., Burns, K. H., Gehring, M., Gorbunova, V., Seluanov, A., & Hammell, M. (2018). Ten things you should know about transposable elements. Genome Biology, 19(1), 199.
- Feschotte, C., & Pritham, E. J. (2007). DNA transposons and the evolution of eukaryotic genomes. Annual Review of Genetics, 41, 331-368.
- Cordaux, R., & Batzer, M. A. (2009). The impact of retrotransposons on human genome evolution. Nature Reviews Genetics, 10(10), 691-703.
- Bourque, G. (2009). Transposable elements in gene regulation and in the evolution of vertebrate genomes. Current Opinion in Genetics & Development, 19(6), 607-612.