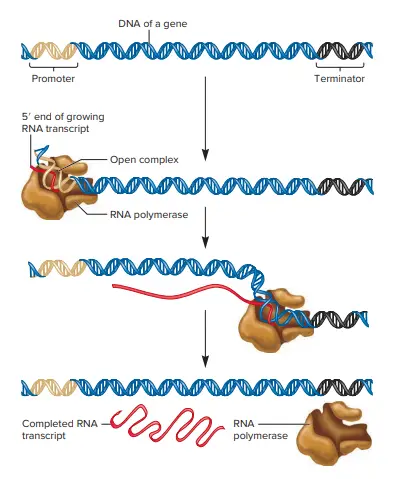
Transcription refers to the process in which the information contained in the DNA strand is transformed into a new messenger RNA molecule (mRNA). In prokaryotic cells, transcription occurs in three stages called the initiation, the elongation, and the termination.
Transcription in prokaryotes
- The essential aspects of transcription are the same for prokaryotes and Eukaryotes, however certain details, like the promoter sequences.
- The RNA polymerase from E. coli has been extensively studied and is the subject of this article. It is the catalyst for all RNA synthesis within this group. The RNA polymerases in archaea have distinct designs; therefore, they will not be discussed here.
- A DNA segment that is transcribed to create one RNA molecule is known as the transcription unit.
- The transcription units could be identical to specific genes, or contain multiple connected genes.
- The large transcripts that contain the coding sequences for several genes are found in bacteria.
- Transcriptional processes could be divided into three steps: (1) initiation of an RNA chain that is not yet in existence, (2) extension of the chain and (3) the end of transcription , and the release of newly formed the RNA molecules.
- When discussing transcription, biologists typically employ the terms upstream and downstream to mean areas that lie towards the 5 and 3rd end, respectively, of the transcript that originates from a area of the mRNA. These terms stem from being aware that RNA synthesizing is always in the 5-3 direction. The regions downstream and upstream of genes contain DNA sequences that define the five and three segments in their transcription in relation to a particular reference location.
Stages of Prokaryotic Transcription
The transcription process takes place in three steps that are initiation, elongation and synthesis transcript; and finally, termination
- Initiation: The Promoter serves as a recognition spot to recognize transcription factor (not illustrated). The transcription factors permit the RNA polymerase to attach to the promoter. After binding, DNA is denatured to form an open complex. This is called the open complex.
- Elongation/synthesis of the RNA transcript: The RNA polymerase slides across the DNA within an open structure to produce the RNA.
- Termination: When a terminator has been achieved that causes an RNA polymerase, as well as the transcript of RNA to break away from DNA.
What are Rna Polymerases: Complex Enzymes?
- The RNA polymerases that trigger transcription are multimeric, complex proteins.
- The E. coli polymerase has a molecular mass of approximately 480,000 and is composed of 5 polypeptides.
- Two of them are identical and, consequently, the enzyme has four polypeptides that are distinct. The entire RNA polymerase molecule is a holoenzyme. It is composed of α2ββ’σ.
- The a subunits participate in the formation of the core tetrameric (α2ββ’) of RNA polymerase.
- The b subunit houses the ribonucleoside-triphosphate binding site as well as the subunit β’ is home to the DNA template binding region.
- One subunit known as the Sigma (σ) factor is only involved in the process of initiation of transcription. It plays not a role in the elongation of chains. Once RNA chain initiation has been achieved, the s factor is released and chain lengthening is catalyzed the main enzyme (α2ββ’).
- Sigma’s role is to identify and bind RNA polymerase to transcription initiation or promoter sites within DNA.
- The enzyme that is the core (with the absence of σ) is able to catalyze RNA synthesis by using DNA template in vitro however, while doing so it will start RNA chains at random locations on both DNA strands. The homoenzyme (σ present) creates RNA chains in vitro but only on sites in live.
Initiation Of RNA Chains/Initiation Of Prokaryotic Transcription
Initiation of RNA chains requires three steps:
- The RNA polymerase binds the homoenzyme to the promoter region in DNA.
- the localized unwinding of two DNA strands through the RNA polymerase. It provides the template strand that is that is free to base-pair with the incoming Ribonucleotides.
- it is the process of creating phosphodiester bond between the initial ribonucleotides of the newly formed DNA chain.
- The holoenzyme stays in the promoter zone during synthesizing of the initial 8 or 9 bonds.
- after which the sigma factor gets released, and the enzyme starts the elongation phase of the synthesis of RNA.
- At the time of initiation short chains of two to nine ribonucleotides are created and released.
- The abortive synthesis ceases after chains that contain 10 or more ribonucleotides have been synthesized . the RNA polymerase is beginning to migrate away of the promoter.
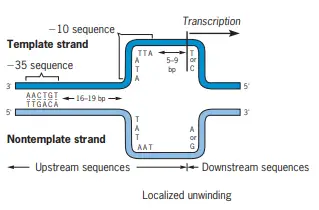
- According to convention it is the case that the nucleotide pair or nucleotides in and adjacent the transcription unit are identified in relation to the transcription beginning location (designated +1)–the nucleotide pair that corresponds to the initial (5′) nucleotide of the transcript of RNA.
- Base pairs prior to the initiation site are identified with without (+) prefixes. Those following (relative in direction to) the site’s initiation are identified with (+) prefixes.
- Nucleotide sequences that precede the site of initiation are known as upstream sequences. Sequences following the initiation sites are known as downstream sequences. As we mentioned previously the sigma subunit in the RNA polymerase binds to DNA promoters.
- Many E. coli promoters have been sequenced and have been found to have very few similarities.
- Two short sequences inside these promoters are well conserved to be recognized. However, even they’re not identical between two promoters.
- The midpoints of the two conserved sequences occur at about 10 and 35 nucleotide pairs, respectively, before the transcription-initiation site. They are referred to as”the 10 sequence” and “the 35 sequence respectively.
- While these sequences differ slightly between genes, certain nucleotides are extremely conserved. The sequences of nucleotides found in these genetic elements are typically known as consensus sequences.
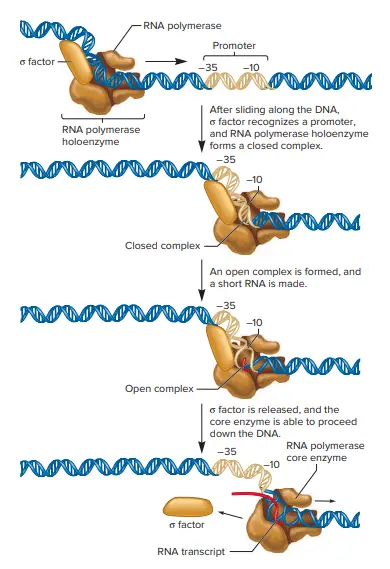
- The consensus sequence for the 10 in the nontemplate strand TATAAT. The 35 consensus sequence, TTGACA.
- The sigma subunit first recognizes and connects with the sequence 35 therefore, the sequence is often referred to as”the recognition sequence.
- This AT-enriched 10 sequence aids in the localized unwinding and unwinding of DNA which is the primary element in the formation of the RNA chain.
- This distance is preserved for E. coli promoters, not shorter than 15 or more than 20 nucleotide pair in length. Additionally, the first or 5th base of E. coli RNAs is generally (90 percent) purine.
Elongation Of RNA Chains
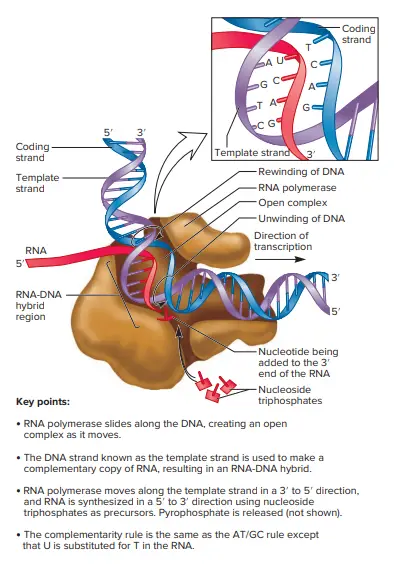
- The lengthening of RNA chains is caused by the RNA Polymerase core enzyme after it has released the subunit. The expansion of the RNA chain occurs in the transcription bubble an unwound region of DNA.
- The RNA polymerase molecules contain DNA unwinding and Rewinding functions.
- RNA polymerase is constantly unwinding the double helix DNA in front of the polymerization site. It also is able to rewind the DNA strands in the background of the polymerization site while it travels across the double Helix.
- The study of E. coli, the average length of a bubble in transcription is about 18 nucleotide pairs approximately 40 ribonucleotides get integrated into the growing RNA chain every second.
- The newly formed RNA chain is displaces by the template DNA strand when the RNA polymerase travels through its DNA-molecule.
- The transient area of base-pairing that occurs between the growing chain with the DNA template is extremely small, possibly only three base pairs.
- It is ensured principally by the binding of DNA and the chain of RNA to the RNA polymerase, not by the binding of DNA’s template strand and the newly formed RNA.
Termination Phase of Prokaryotic Transcription
- The final stage of RNA synthesis is often referred to as the termination.
- Prior to the termination the hydrogen bonding that occurs between DNA and RNA in the open complex is of crucial importance in preventing the separation of RNA polymerase from its template strand.
- The process of termination occurs when the short DNA-RNA hybrid region is made to break apart, thus releasing RNA polymerase and the newly produced RNA transcript.
- Within E. coli, two different ways to terminate transcription have been discovered. For some genes, an RNA-binding protein called “ρ” (rho) has been identified as the one responsible for halting transcription through a process known as the ρ-dependent termination.
- In the case of different genes, terminating doesn’t necessitate the participation of the ρ protein. in these instances the process is referred to as an ρ-independent termination.
ρ-Dependent Termination of Prokaryotic Transcription
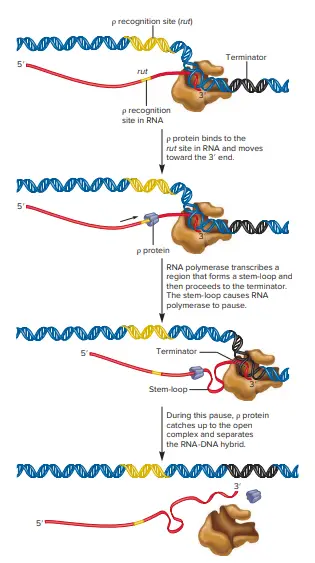
- In the case of ρ-dependent termination the process of termination requires two parts. A first site within the DNA, referred to as the Rut site (for the utilization site for rho) contains an RNA sequence which acts as a recognition spot for binding to the protein ρ.
- What is the role of r protein in the process of termination? The r protein acts as a helicase enzyme that is able to separate RNA and DNA hybrid regions.
- Once the rut site has been produced in the transcript, ρ protein is bound to the RNA, and then moves along the direction of the RNA polymerase.
- The second aspect of ρ-dependent termination is that actually causes termination. This is the site where termination takes place. DNA coded by DNA is an RNA sequence with numerous GC base pairs, which form the stem-loop shape. The process of synthesis of RNA ends with a number of nucleotides that extend beyond the stem-loop. A stem-loop also known as hairpin, may form due to the complementary sequences in the RNA. The stem-loop develops shortly after the RNA sequence has been synthesized and is quickly bound to the RNA polymerase.
- The binding causes an alteration in the conformation that results in a conformational change that causes RNA polymerase to stop its creation of RNA. This pause permits r protein to reach the stem-loop, move over it and rupture hydrogen bonds between DNA and RNA in the complex that is open. If this happens, the RNA strand has been separate from DNA with the RNA polymerase.
ρ-Independent Termination of Prokaryotic Transcription
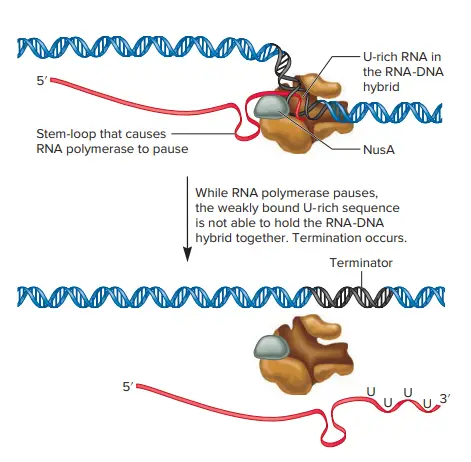
- This process is called ρ-independent terminator is not dependent on an ρ protein. In this instance the terminator consists of two distinct nucleotide sequences.
- One sequence encourages the formation of an a stem-loop. A second one, just downstream of the stem-loop is a uracil rich sequence found at the 3′ side of the RNA. The stem-loop’s formation triggers RNA polymerase to stop its production of RNA.
- This pause will be stabilized through other proteins that attach to the RNA polymerase. For instance the protein NusA is a RNA polymerase binder and helps pausing stem-loop sequences.
- When RNA polymerase is paused it, the uracil-rich sequence of the RNA transcript is bonded with the template DNA. As we have mentioned earlier the hydrogen bonding of DNA to RNA keeps the RNA polymerase in place on DNA.
- However, the uracil-rich pattern to DNA’s template is weak, which causes the RNA transcript to automatically separate from DNA, and stop any further transcription.
- Since this process doesn’t require the protein r to physically eliminate the RNA transcript out of the DNA, it’s also known as intrinsic termination. Within E. coli, about half of genes exhibit intrinsic termination. The majority of them are terminated through the r protein.
RNA processing
- In prokaryotes, RNA transcription from proteins-coding genes (messenger RNA, also known as mRNA) is not a requirement and requires any modification in order to translate.
- Many mRNA molecules start to be translated before RNA synthesizing has been completed.
- However, as ribosomal RNA (rRNA) as well as transfer RNA (tRNA) are made in the form of precursor molecules, both are subject to post-transcriptional processing.
Significance of Transcription
- The transcription of DNA can be described as the technique to regulate the expression of genes.
- It is present in the process of preparation for and is essential to translate proteins.
FAQ
What is the initiation factor during transcription in prokaryotes?
In prokaryotes, translation initiation is controlled by three initiation factors: IF1, IF2, and IF3. Both IF1 and IF2 are involved in positioning the initiator tRNA in the partial P site of the 30S subunit, while the GTPase activity of IF2 signals the beginning of translation elongation (22).
Which one is the termination factor during transcription in prokaryotes?
Transcription termination in prokaryotes can be rho-independent (intrinsic terminators exist in the RNA polymerase) and rho-dependent, i.e., the RNA polymerase requires the cofactor rho for termination of transcription.
What are the steps of transcription in prokaryotes?
Step 1. Initiation
Initialization is the first step in transcription. It happens in the moment that an enzyme called RNA Polymerase connects to a specific area of a gene referred to as”the promoter. The DNA is then triggered to break up so that it can read the bases on some of the DNA chains. It is then able to form the strand of mRNA that has an encapsulated pattern of bases.
Step 2: Elongation
The process of elongation refers to the increase of nucleotides the MRNA strand. The RNA polymerase scans the wound DNA strand and creates the mRNA molecules making use of complementary base pairs. In this process an Adenine (A) in DNA bonds to an Uracil (U) within the RNA.
Step 3. Termination
The termination is the termination of transcription. It happens when the RNA polymerase cross-walks the end (termination) segment within the gene. The mRNA strand has been completed and separates from DNA.