James Wang discovered the first DNA topoisomerase in bacteria in 1971, initially naming it ω (omega) protein; it is now known as Escherichia coli (E. coli) topoisomerase I (topo I) and is a member of the type IA family of enzymes. Subsequently, James Champoux and Renato Dulbecco discovered an analogous activity in eukaryotic cells (rat liver); the enzyme responsible, eukaryotic topo I, has a distinct mechanism and is representative of the type IB family. DNA gyrase from bacteria was the first type II topoisomerase to be discovered, discovered by Martin Gellert and colleagues in 1976 and characterized by Nicholas Cozzarelli and colleagues.DNA gyrase is the only type II enzyme that catalyzes the incorporation of negative supercoils into DNA; all others catalyze DNA relaxation. Type II enzymes are mechanistically distinct from type I enzymes in that they are ATP-dependent and transiently cleave both DNA strands. Subsequently, topoisomerases of type II were isolated from bacteria, viruses, and eukaryotes. Here are the Topo EC-codes: ATP-independent (type I), EC 5.6.2.1; ATP-dependent (type II): EC 5.6.2.2. The only exception among type I topoisomerases is reverse gyrase, which contains a helicase domain (EC 3.6.4.12) and induces ATP-dependent positive supercoiling. Consequently, it is the only topoisomerase of type I classified as EC 5.6.2.2.
What is Topoisomerase?
- Topoisomerases are essential enzymes that play a crucial role in various processes involving DNA. They were first discovered by J.C. Wang in the 1970s while studying Escherichia coli, specifically identifying the type I topoisomerase.
- The primary function of topoisomerases is to modify the topology of DNA, meaning they can change the structural arrangement of the DNA molecule. They have the ability to increase or decrease the degree of DNA unwinding, which is important for processes such as DNA replication, chromosome segregation, transcription, and recombination.
- Topoisomerases are often referred to as DNA topoisomerases because they specifically act on DNA strands and not on RNA. They accomplish their tasks by breaking the phosphodiester bonds that hold the DNA backbone together. However, these breaks are transient and reversible. Once the necessary changes have been made, the enzyme reseals the DNA backbone by reforming the phosphodiester bonds.
- There are two major types of DNA topoisomerases: type I and type II. Type I topoisomerases cleave only one DNA strand, while type II topoisomerases cleave both strands of the DNA molecule. By cutting the DNA strands, these enzymes can relieve the torsional stress and resolve topological problems associated with DNA overwinding, tangling, and knotting.
- The presence of topological issues in DNA can hinder crucial biological processes. For instance, overwinding of the DNA duplex during DNA replication or transcription can impede the progress of DNA or RNA polymerases. Moreover, the linking or tangling of DNA strands during replication can interfere with cell division. DNA topoisomerases act as guardians against such complications by intervening, cutting the DNA strands, and allowing the necessary untangling, unwinding, or unknotting to take place. Once the process is complete, the DNA strands are rejoined, and the original topology of the DNA is restored.
- In summary, topoisomerases are enzymes that modify the topological state of DNA by temporarily breaking and resealing the DNA backbone. They play a critical role in DNA replication, transcription, chromosome segregation, and recombination by resolving topological problems that can impede these processes. By understanding and harnessing the functions of topoisomerases, researchers can gain insights into DNA dynamics and develop strategies to target these enzymes for therapeutic purposes.
In the realm of topoisomerases, several important terms help describe the structural characteristics and properties of DNA.
One such term is “twist” (Tw), which refers to the total count of helical turns made by the individual strands of DNA. The twist is a measure of the winding of the DNA double helix.
Another crucial term is “writhe” (Wr), which represents the total count of turns made by the double helix of DNA as it crosses over itself. Writhe is associated with the supercoiling of DNA and indicates the extent to which the DNA molecule is twisted upon itself.
The “linking number” (Lk) is a fundamental concept in topoisomerases, and it encompasses both the twist and writhe of DNA. The linking number is determined by adding the twist and writhe values together.
Mathematically, the linking number can be expressed as Lk = Wr + Tw, where Wr represents the writhe and Tw represents the twist. By quantifying the linking number, scientists can gain insights into the overall topology and supercoiling status of DNA.
These terms and their interplay play a significant role in understanding the actions of topoisomerases. By manipulating the twist and writhe of DNA, these enzymes can modify the linking number and alleviate the topological constraints that may impede critical DNA processes such as replication, transcription, and chromosome segregation.
Topoisomerase Definition
Topoisomerase is an enzyme that modifies the topology of DNA by cutting and rejoining the DNA strands, allowing for untangling, unwinding, or unknotting of the DNA molecule.
Types of Topoisomerase
There are two major types of topoisomerases: type I and type II. Here’s a brief overview of each type:
- Type I Topoisomerases (Topoisomerase i): Type I topoisomerases catalyze changes in DNA topology by cleaving one strand of the DNA molecule. This break allows for the relaxation of DNA supercoiling, the removal of knots or tangles, and the resolution of topological problems. After the necessary changes have been made, the enzyme reseals the DNA strand, restoring the original DNA structure. Type I topoisomerases are further divided into two subtypes: IA and IB.
- Type IA Topoisomerases: Type IA topoisomerases introduce transient single-stranded breaks in the DNA and can change the linking number by increments of one.
- Type IB Topoisomerases: Type IB topoisomerases also create transient single-stranded breaks in the DNA but do not require ATP hydrolysis. They can change the linking number by increments of one.
- Type II Topoisomerases (Topoisomerase ii): Type II topoisomerases are ATP-dependent enzymes that break and rejoin both strands of the DNA molecule. These enzymes are capable of resolving more complex topological problems, such as catenations (linked DNA rings) and knots. Type II topoisomerases can change the linking number by increments of two. They are further divided into two subtypes: IIA and IIB.
- Type IIA Topoisomerases: Type IIA topoisomerases form a transient covalent linkage with the DNA during the cleavage and rejoining process. This covalent intermediate is referred to as a “cleavage complex.” Examples of type IIA topoisomerases include bacterial DNA gyrase and eukaryotic topoisomerase II (topo II).
- Type IIB Topoisomerases: Type IIB topoisomerases do not form a covalent linkage with the DNA during their catalytic cycle. They play a crucial role in DNA replication, chromosome segregation, and DNA repair processes. Topoisomerase VI is an example of a type IIB topoisomerase.
Both type I and type II topoisomerases are essential for maintaining DNA structure, resolving topological problems, and enabling various DNA-dependent processes in cells. Their distinct mechanisms and activities make them critical players in DNA metabolism and regulation.
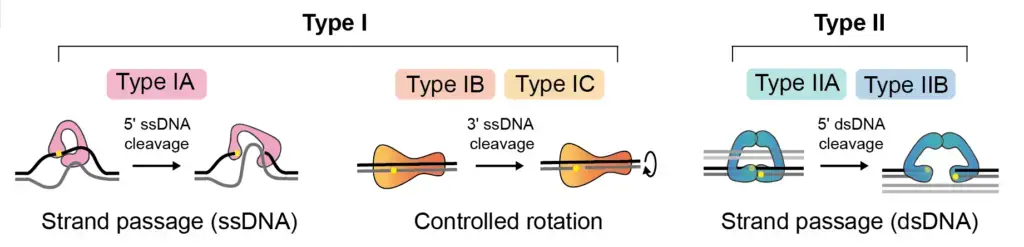
Type I Topoisomerase (Topoisomerase i/Topoisomerase 1)
Definition of Type I Topoisomerase
- Type I topoisomerases are a type of topoisomerase enzyme that play a crucial role in altering the topology of DNA. Unlike type II topoisomerases, they cleave a single strand of DNA during their catalytic cycle. It is important to note that although they are called “type I,” they are not ATP-dependent enzymes, except for a unique exception known as Reverse Gyrase.
- The primary function of type I topoisomerases is to change the linking number of DNA by incrementing it by one. They achieve this by creating transient breaks in the DNA backbone, allowing the DNA to undergo relaxation, unknotting, and decatenation processes. These enzymes can act on both single-stranded and double-stranded DNA substrates, and they can employ different mechanisms such as a “swivel” or “strand-passage” mechanism to facilitate their reactions.
- The reactions catalyzed by type I topoisomerases include DNA supercoil relaxation, the unknotting of single-stranded circular DNA, and decatenation. However, it’s important to note that decatenation requires at least one partner DNA molecule to have a single-stranded region. It is also worth mentioning that a unique type I topoisomerase called reverse gyrase, found in archaea, is capable of introducing positive supercoiling in DNA.
- In summary, type I topoisomerases are a subtype of topoisomerases that cleave a single strand of DNA and play a significant role in changing the linking number of DNA by plus one. They can relax DNA supercoils, untangle circular DNA molecules, and decatenate DNA strands, provided the necessary single-stranded regions are present. However, it is important to remember that reverse gyrase, an exception within type I topoisomerases, is an ATP-dependent enzyme capable of introducing positive supercoiling in DNA.
Structure of Type I Topoisomerase
- The structure of type I topoisomerases can vary depending on the subtype, with type IA and type IB having distinct characteristics.
- In type IA topoisomerases, there are multiple domains that can range from I to IV. The Toprim domain is located within domain I, which is involved in DNA binding and cleavage activity. Domains III and IV contain the Helix-Turn-Helix (HTH) motif, which is a DNA-binding motif. Specifically, in domain III, there are tyrosine residues that are important for the catalytic activity of the enzyme. When observing the overall structure, type IA topoisomerases can resemble a lock, with all three domains located at the bottom of the topoisomerase structure.
- Type IB topoisomerases, on the other hand, have a different structure. They possess an active site, typically a tyrosine residue, which is involved in DNA cleavage and rejoining. Type IB topoisomerases consist of several domains, including the C-terminal domain, N-terminal domain, capping, and catalytic lobe. These domains work together to facilitate the binding of DNA and the catalytic activities of the enzyme.
- Overall, the structure of type I topoisomerases, whether type IA or type IB, is composed of multiple domains that are essential for their DNA-binding and enzymatic functions. The specific arrangement and interactions between these domains contribute to the overall functionality and mechanism of the type I topoisomerase enzymes.
Type I Topoisomerase Types
Type I topoisomerases are categorized into three basic types based on their characteristics and binding sites:
- Type IA Topoisomerases: Type IA topoisomerases bind to the 5′ carbon end of the DNA molecule. They share homology with the topoisomerase I enzyme found in Escherichia coli. Type IA topoisomerases can be further divided into three subtypes:
- Topo IA: This subtype is found in eubacteria. It plays a role in DNA topology changes within bacterial cells.
- Topo III: Topo III is present in both eubacteria and eukaryotes. It participates in various DNA processes, including recombination, DNA repair, and chromosome segregation.
- Reverse Gyrase: Reverse gyrase is a unique subtype of type IA topoisomerase found in archaebacteria and some eubacteria. It is the only type I topoisomerase that requires ATP for its catalytic activity. Reverse gyrase is capable of introducing positive supercoiling in DNA and is involved in DNA stability at high temperatures.
- Type IB Topoisomerases: Type IB topoisomerases bind to the 3′ carbon end of the DNA molecule and create a nick in one strand. They exhibit homology to the human topoisomerase I enzyme. Type IB topoisomerases are involved in DNA relaxation, replication, and repair processes.
- Type IC Topoisomerases: Type IC topoisomerases consist of a single subtype known as topoisomerase V. They bind to the 3′ carbon end of the DNA molecule and are primarily found in archaebacteria. Type IC topoisomerases exhibit a controlled mechanism of rotation and are involved in various DNA transactions within archaebacterial cells.
Mechanism of action of Type I Topoisomerase
Type IA Mechanims
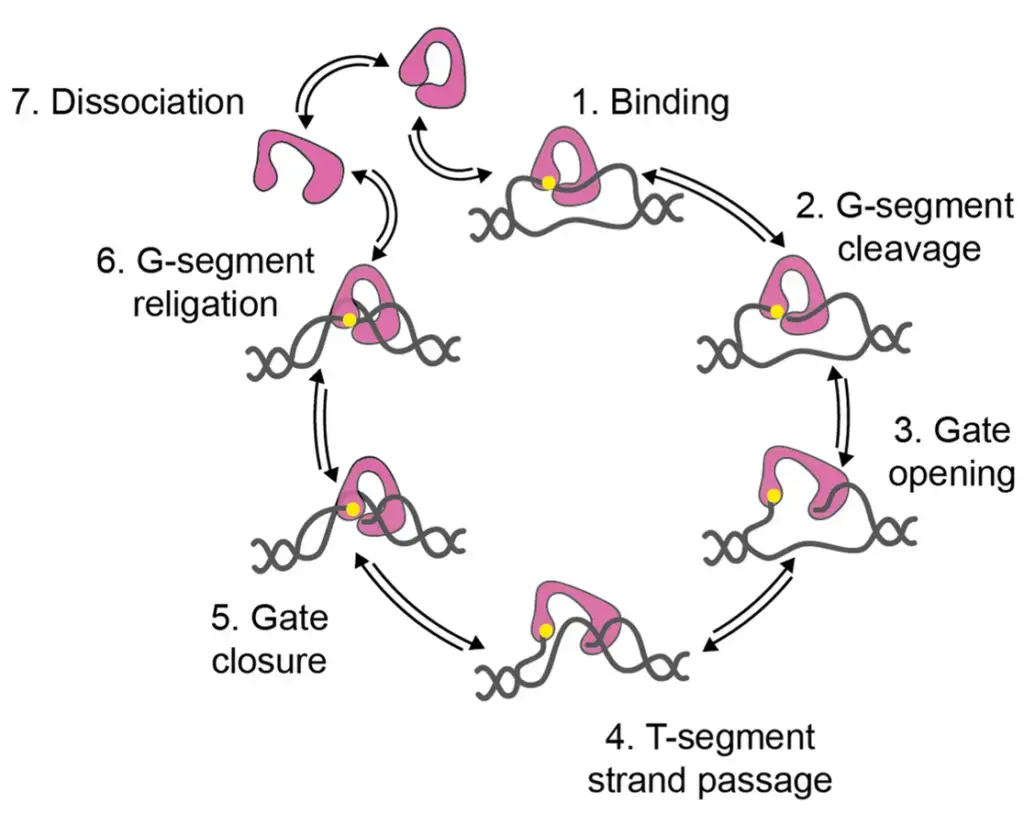
The strand-passage mechanism for type IA topoisomerases involves several steps:
- Topoisomerase binding: The type IA topoisomerase binds to a single-stranded region of DNA known as the G-segment.
- Cleavage of the G-segment: The G-segment of DNA is cleaved by the topoisomerase, creating a transient single-stranded break. This break is facilitated by the formation of a tyrosyl-phosphate bond between a tyrosine residue in the enzyme and a 5′-phosphate in the DNA.
- Opening of the DNA gate: The topoisomerase undergoes a conformational change, opening the DNA gate. This allows for the passage of another segment of DNA called the T-segment through the cleaved G-strand.
- Transfer of the T-segment: The T-segment is transferred through the cleaved G-strand, facilitated by the opened DNA gate.
- Closure of the DNA gate: After the T-segment has been transferred, the DNA gate of the topoisomerase is closed.
- Re-ligation of the G-segment: The cleaved G-strand is re-ligated, restoring the integrity of the DNA molecule. This re-ligation process changes the linking number of the DNA by one.
- Repeat or dissociation: The topoisomerase can undergo another round of relaxation, repeating the strand-passage process, or it can dissociate from the DNA.
Type IA topoisomerases, which are monomeric enzymes, play a role in DNA relaxation. They introduce transient single-stranded breaks in DNA through the formation of tyrosyl-phosphate bonds. The cleavage of the G-segment allows for the passage of the T-segment, followed by re-ligation of the G-segment. This strand-passage process changes the linking number of the DNA by +/-1. Examples of type IA topoisomerases include prokaryotic topoisomerase I and III, eukaryotic topoisomerase IIIα and IIIβ, and the archaeal enzyme reverse gyrase. Reverse gyrase, found in thermophilic archaea, is unique as it can introduce positive supercoils into DNA.
Type IB
The strand-passage mechanism for Type IB topoisomerases differs from that of Type IA topoisomerases and operates through a process known as “controlled rotation” or “swivel” mechanism. Here are the key features of the strand-passage mechanism for Type IB topoisomerases:
- Transient single-stranded breaks: Type IB topoisomerases catalyze reactions involving transient single-stranded breaks in DNA. These breaks are created through the formation of a tyrosyl-phosphate bond between a tyrosine residue in the enzyme and a 3′-phosphate in the DNA.
- Controlled rotation: Unlike the strand-passage mechanism seen in Type IA topoisomerases, Type IB topoisomerases use a controlled rotation mechanism. This mechanism allows the cleaved strand to rotate around the intact strand in a swiveling motion.
- Variable change in linking number: The controlled rotation of the cleaved strand around the intact strand leads to a variable change in the linking number per cleavage and religation event. The extent of rotation and resulting change in linking number depend on the specific reaction and the characteristics of the enzyme.
- Nick re-ligation: After the desired rotation or swiveling of the cleaved strand, the transient single-stranded break is re-ligated. This restores the integrity of the DNA molecule.
- Structural and evolutionary distinction: The strand-passage mechanism employed by Type IB topoisomerases is distinct from the mechanism used by Type IA topoisomerases. These two groups of enzymes are structurally and evolutionarily unrelated, despite both being involved in DNA relaxation through the creation of transient breaks.
Type IB topoisomerases, such as eukaryotic nuclear and mitochondrial topo I and viral topo I, utilize the controlled rotation mechanism for their catalytic activities. They are found in all three domains of life (Archaea, Bacteria, and Eukarya) and play important roles in DNA maintenance and regulation.
Topoisomerase 1 Functions
Topoisomerase 1 (Topo 1) performs several important functions in various biological processes:
- Removal of Supercoils: One of the primary functions of Topo 1 is the removal of supercoils from DNA. Supercoiling occurs when the DNA helix becomes twisted or overwound, and it can impede processes such as replication and transcription. Topo 1 acts by introducing transient breaks in the DNA backbone, allowing the relaxation and removal of supercoils, thereby facilitating proper DNA replication and gene expression.
- DNA Relaxation: Topo 1 plays a crucial role in relaxing DNA. By introducing reversible single-strand breaks in the DNA, it alleviates torsional strain and allows the DNA to unwind, promoting DNA unwinding and facilitating DNA processes such as transcription and replication.
- Strand Breaking during Recombination: During recombination, the exchange of genetic material occurs between DNA molecules. Topo 1 participates in this process by breaking DNA strands, allowing for the exchange and rearrangement of genetic information, and subsequently rejoining the DNA strands.
- Chromosome Condensation: Topo 1 is involved in the condensation of chromosomes, particularly during cell division. It helps in resolving the interwinding and tangling of DNA strands that occur during mitosis, ensuring the proper segregation of chromosomes into daughter cells.
Type II Topoisomerase (Topoisomerase ii/Topoisomerase 2)
Topoisomerase 2 Definition
Type II topoisomerase is a specific type of topoisomerase enzyme that plays a crucial role in DNA topology and regulation. Unlike Type I topoisomerases that cut a single strand of DNA, Type II topoisomerases cleave both strands of DNA simultaneously, allowing for more extensive changes in DNA topology. This process requires the energy provided by ATP hydrolysis, making Type II topoisomerases ATP-dependent enzymes.
One of the key functions of Type II topoisomerases is changing the linking number of DNA by two. The linking number refers to the total number of twists and writhes in the DNA molecule. By introducing a double-stranded break and allowing the DNA to pass through, Type II topoisomerases can alter the linking number by two units. This process is essential for relieving torsional stress, resolving DNA supercoils, and regulating DNA topology.
Type II topoisomerases are involved in a wide range of cellular processes, including DNA replication, transcription, recombination, and chromosome condensation. They play a critical role in separating and untangling replicated DNA strands, ensuring proper chromosome segregation during cell division. Moreover, they are necessary for DNA packaging, condensing the long DNA molecules into compact and organized structures.
Here are the key features of Type II topoisomerases:
- Reactions on double-stranded DNA: Type II topoisomerases act on double-stranded DNA substrates and facilitate various reactions, including DNA relaxation, DNA supercoiling, unknotting, and decatenation. These enzymes have a wide range of functions related to DNA topology.
- Strand-passage mechanism: Type II topoisomerases employ a strand-passage mechanism during their catalytic activities. The process involves the binding of one DNA duplex, known as the gate segment (G-segment), at the DNA gate. Another duplex, called the transport segment (T-segment), is captured by an ATP-operated clamp. The T-segment is then passed through a transient break in the G-segment, which involves the formation of 5ʹ phosphotyrosine linkages in both DNA strands. Finally, the T-segment is released through the C-gate, and the G-segment is re-ligated.
- Dimeric or tetrameric structure: Type II topoisomerases are generally found as homodimers or heterotetramers, in contrast to the monomeric nature of type I topoisomerases. The specific structure of these enzymes allows for the coordination of multiple DNA strands and the proper functioning of the strand-passage mechanism.
- Subtypes: Type II topoisomerases are classified into two subtypes based on evolutionary, structural, and mechanistic considerations. These subtypes exhibit variations in their functions and mechanisms of action.
- ATP-dependent turnover: Enzyme turnover in Type II topoisomerases is dependent on the binding and hydrolysis of ATP. ATP provides the necessary energy for the conformational changes and movements involved in the strand-passage mechanism.
One prominent example of a Type II topoisomerase is gyrase, a bacterial enzyme that can introduce negative supercoils in addition to catalyzing DNA relaxation. Type II topoisomerases are essential for various cellular processes, including DNA replication, transcription, and chromosome condensation. Their ability to modulate DNA topology is crucial for maintaining proper DNA structure and function.
Topoisomerase 2 Structure
The structure of Type II topoisomerases, specifically Topoisomerase IIA and Topoisomerase IIB, can vary between eukaryotes and prokaryotes:
- Topoisomerase IIA: In eukaryotes, Topoisomerase IIA is composed of two identical monomers, forming a dimeric structure (A-A). However, in prokaryotes, it forms a heterotetrameric structure with two different subunits (A2B2).
- Topoisomerase IIB: Topoisomerase IIB is exclusively formed by heterotetramers, consisting of two different subunits.
The overall structure of Type II topoisomerases typically includes four distinct domains:
- ATPase Domain: Located at the N-terminal region, the ATPase domain is responsible for the ATP-dependent activity of Type II topoisomerases. It hydrolyzes ATP to provide the energy required for the enzymatic functions of these topoisomerases.
- C-Terminal Domain: The C-terminal domain of Type II topoisomerases is variable and can vary in size and composition. This region often plays a regulatory role and can be involved in protein-protein interactions or binding to specific targets.
- DNA Binding Domain: Positioned in the central region of the enzyme, the DNA binding domain is responsible for the interaction with DNA molecules. It enables the recognition and binding of specific DNA sequences, facilitating the cleavage and rejoining of DNA strands.
- Toprim Domain: The Toprim domain is a conserved domain of approximately one hundred amino acids. It is involved in the catalytic activity of Type II topoisomerases and plays a crucial role in the DNA cleavage and religation processes.
These structural features allow Type II topoisomerases to perform their functions, including DNA strand cleavage, passage, and rejoining. The ATPase domain provides the energy necessary for these enzymatic activities, while the DNA binding domain and Toprim domain play key roles in DNA interaction and catalysis.
Overall, the structure of Type II topoisomerases consists of multiple domains that work together to enable their essential functions in DNA topology regulation and various cellular processes.
Types of Topoisomerase 2
Type II topoisomerases can be categorized into two main types:
- Type IIA Topoisomerases: Type IIA topoisomerases are found in viruses and all cellular organisms. This type includes three subtypes:
- Topo II: Topo II is primarily found in eukaryotes. It plays a critical role in various DNA processes, including DNA replication, transcription, and chromosome segregation. Topo II is involved in the regulation of DNA topology by introducing reversible double-stranded breaks and allowing DNA passage through these breaks.
- Topo IV: Topo IV is primarily found in bacteria. It differs from Gyrase in its function. While Gyrase is involved in DNA wrapping and promoting negative supercoiling, Topo IV is not involved in DNA wrapping. Topo IV is crucial for bacterial DNA segregation during cell division and helps resolve interlinked DNA strands.
- Gyrase: Gyrase is found in bacteria and some eukaryotes. It is a type II topoisomerase that introduces negative supercoiling into DNA. By decreasing the linking number by two, gyrase helps maintain the proper level of DNA supercoiling in bacteria, which is essential for DNA replication and gene expression.
- Type IIB Topoisomerases: Type IIB topoisomerases include Topo VI, which is found in archaea and some plants. Topo VI is structurally distinct from Type IIA topoisomerases and has unique functions in DNA metabolism. It plays a role in DNA relaxation, knotting, and unknotting, and is involved in processes such as DNA replication and recombination.
Type IIA features
Type IIA topoisomerases play a crucial role in catalyzing transient double-stranded breaks in DNA and are involved in various DNA topology changes. Here are the key features and characteristics of Type IIA topoisomerases:
- Mechanism of action: Type IIA topoisomerases catalyze the formation of tyrosyl-phosphate bonds between tyrosine residues in the enzyme and 5′-phosphates in DNA strands. This results in transient double-stranded breaks in the DNA.
- Strand-passage reaction: Type IIA topoisomerases can facilitate both intra- and intermolecular strand-passage reactions. In the intra-molecular reaction, changes in supercoiling and knotting of the DNA molecule can occur. In the intermolecular reaction, the enzyme is involved in unlinking DNA strands.
- Linking number changes: The strand-passage reaction mediated by Type IIA topoisomerases alters the linking number of DNA by +/-2, reflecting the changes in DNA supercoiling or unlinking.
- Examples of Type IIA topoisomerases: Eukaryotic topo IIα and topo IIβ, bacterial gyrase, and topo IV are all examples of Type IIA topoisomerases. Gyrase, a bacterial enzyme, is unique as it can introduce negative supercoils into DNA.
- Gyrase-specific features: DNA gyrase, a type of Type IIA topoisomerase, shares the same strand-passage mechanism as other Type IIA enzymes. However, gyrase has distinct characteristics related to its ability to induce negative supercoiling in DNA. It wraps a much longer segment of DNA (over 100 base pairs) around the enzyme, with one arm forming the T-segment that is passed through the double-stranded break.
- ATP-dependent activity: Type IIA topoisomerases, including gyrase, require ATP hydrolysis to perform their functions. The energy from ATP hydrolysis is transduced into torsional stress in DNA, which is essential for processes such as supercoiling.
- ATP-independent activity: In the absence of ATP, gyrase is still capable of performing a slower DNA relaxation reaction, allowing the removal of negative supercoils.
Type IIA topoisomerases, including gyrase, are essential for regulating DNA topology and are involved in DNA replication, transcription, and other cellular processes. Their ability to introduce and remove supercoils in DNA is crucial for maintaining proper DNA structure and function.
Type IIB features
Type IIB topoisomerases are a distinct subgroup of topoisomerases that catalyze transient double-stranded breaks in DNA, similar to Type IIA enzymes. Here are the key features and characteristics of Type IIB topoisomerases:
- Mechanism of action: Type IIB topoisomerases catalyze the formation of tyrosyl-phosphate bonds between tyrosine residues in the enzyme and 5′-phosphates in opposite strands of DNA. This results in transient double-stranded breaks in the DNA. However, in Type IIB enzymes, the double-stranded breaks have a 2-base stagger, meaning the breaks occur at specific positions with a two-base offset.
- Structural differences: Type IIB topoisomerases exhibit important structural differences compared to Type IIA enzymes. One notable difference is the absence of one of the protein “gates,” known as the C gate. The C gate is involved in the strand-passage mechanism of Type IIA topoisomerases but is not present in Type IIB enzymes.
- Evolutionary relationship: Although Type IIB topoisomerases have distinct structural features, they are evolutionarily related to Type IIA enzymes. Despite their differences, both subgroups share a common ancestry and similar catalytic mechanisms.
- Distribution: Type IIB topoisomerases were initially discovered in archaea but have also been found in eukaryotes, particularly in plants. Examples of Type IIB topoisomerases include topo VI and topo VIII, with topo VI being the most extensively studied enzyme in this subgroup.
- Preferential decatenase: Topo VI, the well-studied enzyme of the Type IIB sub-type, is believed to function as a preferential decatenase. It is involved in untangling or separating intertwined DNA molecules, known as catenanes, which can occur during DNA replication or recombination processes.
Type IIB topoisomerases, such as topo VI and topo VIII, play important roles in DNA topology regulation, particularly in resolving DNA catenanes. Their ability to catalyze transient double-stranded breaks with a specific 2-base stagger provides them with unique capabilities in maintaining DNA structure and function.
In summary, Type II topoisomerases are divided into Type IIA and Type IIB, each with distinct subtypes. Type IIA includes Topo II, Topo IV, and Gyrase, which are found in different organisms and have specific functions in DNA topology regulation. Type IIB consists of Topo VI, which has unique roles in DNA metabolism and can be found in archaea and some plants.
Mechanism of action of Topoisomerase 2
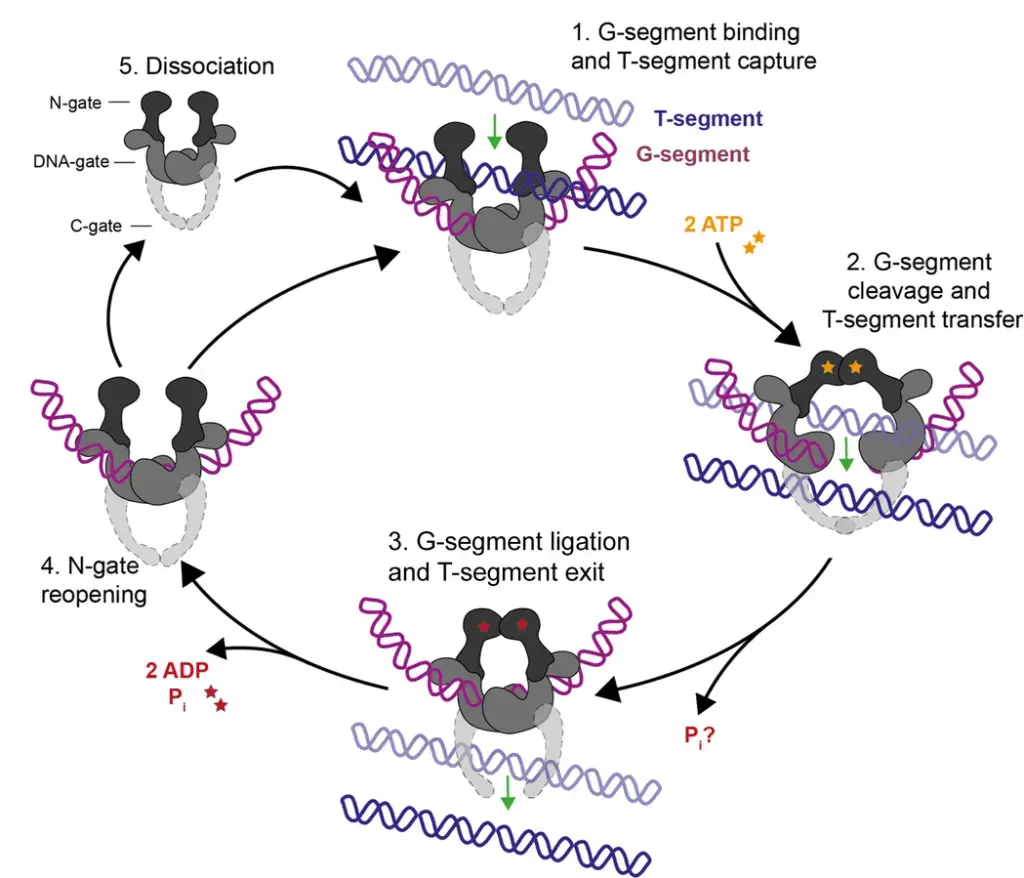
The strand-passage mechanism of Type II topoisomerases involves several steps that allow for the manipulation of DNA topology. Here is an overview of the process:
- Binding of G-segment and capturing the T-segment: The G-segment, a DNA duplex, is initially bound at the DNA-gate of the Type II topoisomerase. Simultaneously, the T-segment, another DNA duplex, is captured by the enzyme.
- ATP binding and dimerization: The binding of ATP stimulates the dimerization of the N-gate, a structural element of the topoisomerase. This conformational change prepares the enzyme for the subsequent steps of the mechanism.
- Cleavage and passage of the T-segment: As the N-gate dimerizes, the G-segment is cleaved, creating a transient double-strand break. This break allows the T-segment to pass through the gap. For Type IIB topoisomerases, which lack a C-gate, the T-segment is released from the enzyme once it has passed through the G-segment.
- Re-ligation of the G-segment: After the T-segment has successfully traversed the G-segment, the double-strand break in the G-segment is re-ligated. This step restores the integrity of the DNA duplex.
- N-gate opening and enzyme dissociation: The dissociation of ADP and phosphate (Pi) from the enzyme allows the N-gate to open. At this point, the enzyme can remain bound to the G-segment, ready to capture another T-segment for further strand-passage cycles. Alternatively, the enzyme may dissociate from the G-segment, completing its role in the current process.
The strand-passage mechanism of Type II topoisomerases requires ATP binding and hydrolysis to drive the conformational changes and energy-dependent steps. Through this mechanism, Type II topoisomerases play essential roles in DNA replication, transcription, and other processes that involve the manipulation of DNA topology.
Topoisomerase 2 Function
Topoisomerase II, also known as DNA gyrase, plays several important functions in DNA metabolism. Here are the key functions associated with Topoisomerase II:
- Disentanglement of Chromosomes: Topoisomerase II plays a crucial role in the disentanglement of chromosomes. During DNA replication and transcription, DNA strands become intertwined, forming complex structures that need to be resolved for proper DNA functioning. Topoisomerase II is responsible for untangling these interlinked DNA strands, ensuring smooth DNA replication, transcription, and other cellular processes.
- Relaxation of DNA: Topoisomerase II is involved in the relaxation of DNA. It helps to relieve DNA supercoiling, which occurs when the double helix of DNA becomes twisted or overwound. By introducing reversible double-stranded breaks in DNA and allowing the passage of DNA strands through these breaks, Topoisomerase II helps to reduce excessive supercoiling, maintaining the appropriate level of DNA tension and ensuring proper DNA function.
- Promotion of Negative Supercoiling: DNA gyrase, a type of Topoisomerase II found in bacteria and some eukaryotes, specifically promotes negative supercoiling of DNA. Negative supercoiling involves the introduction of additional twists in the opposite direction to the natural helical twist of DNA. DNA gyrase utilizes energy from ATP hydrolysis to introduce negative supercoils into DNA, playing a crucial role in DNA compaction and regulation of gene expression in bacteria.
- Change in Linking Number: One of the essential functions of Topoisomerase II is the alteration of the linking number of DNA loops. The linking number represents the total number of times DNA strands are intertwined or linked in a closed circular DNA molecule. Topoisomerase II can change the linking number by two units, either increasing or decreasing it, depending on the specific DNA topology and cellular requirements. This capability allows Topoisomerase II to resolve DNA knots, tangles, and other topological complexities in DNA molecules.
Topoisomerase As drug targets
Topoisomerases are crucial enzymes involved in DNA replication, transcription, and other DNA metabolic processes. Their ability to create transient breaks in DNA makes them attractive targets for drug development, both in antibacterial and anti-cancer chemotherapy. Several drugs that target topoisomerases have been included in the World Health Organization Model List of essential Medicines. Here is an overview of the drugs targeting topoisomerases for antibacterial and anti-cancer purposes:
- Antibacterial compounds: a. Fluoroquinolones (FQs): Fluoroquinolone antibiotics, such as ciprofloxacin, levofloxacin, and moxifloxacin, target bacterial DNA gyrase and DNA topoisomerase IV. These compounds stabilize the DNA-protein covalent cleavage intermediate, preventing DNA religation and causing cell death. However, the emergence of resistance to fluoroquinolones is a significant concern.b. Aminocoumarins: Aminocoumarin compounds like novobiocin, clorobiocin, and coumermycin A1 inhibit the ATPase reaction of gyrase and topoisomerase IV. While they can be potent against their target, issues related to permeability and toxicity have limited their clinical success.c. Proteinaceous inhibitors: Some bacterial toxins, such as CcdB, MccB17, and ParE, stabilize the cleavage complex of gyrase, similar to fluoroquinolones. Although not viable as antibacterials themselves, they serve as a basis for the development of novel antibacterial compounds. Other protein inhibitors, such as YacG and pentapeptide repeat proteins (e.g., QnrB1 and MfpA), prevent DNA binding by the topoisomerase, conferring resistance to fluoroquinolones.
- Anti-cancer compounds: a. Camptothecin (CPT): Derived from the Camptotheca acuminata tree, camptothecin and its derivatives (e.g., topotecan and irinotecan) target human topoisomerase I. They stabilize the topo I cleavage complex, preventing DNA religation and resulting in toxic double-stranded DNA breaks. These compounds are used in chemotherapy for various cancers.b. Etoposide (VP-16): Etoposide and its derivative teniposide are epipodophyllotoxin derivatives that target human topoisomerase II. They stabilize the covalent cleavage complex, preventing DNA religation. These drugs are commonly used in combination with other chemotherapy agents for the treatment of specific cancers.c. Doxorubicin: Doxorubicin and related anthracycline derivatives, including daunorubicin, epirubicin, and idarubicin, target human topoisomerase II. They stabilize the cleavage complex, inhibiting DNA religation. Anthracyclines are widely used in chemotherapy for various cancers, but their use can be limited by cardiotoxicity.d. Other inhibitors: Additional inhibitors, such as merbarone and dexrazoxane, target topoisomerase II but function as catalytic inhibitors rather than stabilizing the cleavage complex. Dexrazoxane is used to prevent cardiotoxicity associated with anthracycline treatment.
These drugs act as topoisomerase poisons by interfering with the DNA-protein cleavage complex, leading to DNA damage and cell death. They have shown efficacy in treating bacterial infections and various types of cancer. However, drug resistance and limitations related to toxicity and stability highlight the ongoing need for the development of new topoisomerase-targeted compounds.
Role of topoisomerase in transcriptional regulation
Topoisomerases play a crucial role in the regulation of gene transcription, and specifically, DNA topoisomerase II beta (topo IIβ) has been identified as having regulatory functions in this process. Topo IIβ is involved in the rapid expression of immediate early genes and signal-responsive gene regulation by facilitating double-strand DNA breaks and interacting with components of the DNA damage repair machinery.
One important aspect of topo IIβ’s role in transcriptional regulation is its involvement in the initiation of transcription. In response to specific stimuli, such as estrogen, serum, insulin, glucocorticoids, and neuronal activation, topo IIβ induces limited, short-term double-strand DNA breaks in the promoter regions of signal-regulated genes. These breaks allow for the rapid up-regulation of gene expression. Once the DNA breaks are repaired, transcription of the signal-responsive gene returns to a low basal level. This mechanism ensures a transient and controlled increase in gene expression in response to external signals.
Topo IIβ acts in conjunction with other associated enzymes to facilitate the release of paused RNA polymerase at highly transcribed or long genes. RNA polymerase II frequently pauses at specific sites located approximately 30-60 nucleotides downstream of the transcription start site of a gene. The pausing of RNA polymerase II and its controlled release are believed to play a regulatory role in gene transcription. Short-term topo IIβ-induced DNA double-strand breaks occur at these pausing sites, and they are necessary for efficient release of the paused state and progression to gene transcription. In fact, around 80% of highly expressed genes in HeLa cells exhibit pausing of RNA polymerase II. Therefore, the DNA double-strand breaks induced by topo IIβ are thought to be part of the regulatory process that controls gene expression.
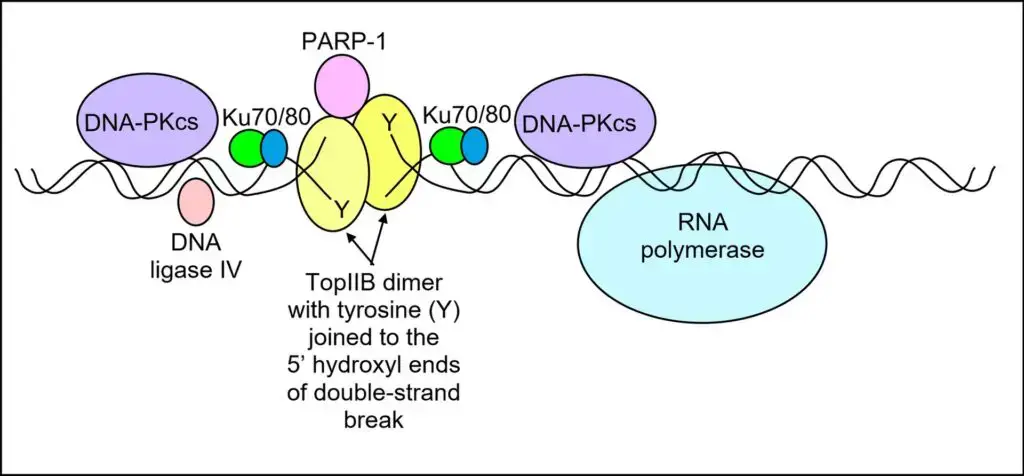
Topo IIβ collaborates with other proteins to execute its role in transcriptional regulation. PARP-1 (poly ADP-ribose polymerase 1) is constitutively present with topo IIβ near the transcription start site of signal-responsive genes. Upon the occurrence of a signal, topo IIβ induces a double-strand DNA break, and PARP-1 replaces histone H1 with HMGB1/HMGA2, which promotes transcription. Additionally, components of the non-homologous end joining DNA repair pathway, including DNA-PKcs, Ku70/Ku80, and DNA ligase IV, assemble with topo IIβ and PARP-1 at the site of the double-strand break. These components are essential for repairing the DNA break and closing the double-stranded DNA break.
In summary, DNA topoisomerase II beta (topo IIβ) plays a regulatory role in gene transcription. It facilitates the rapid expression of immediate early genes and signal-responsive genes by inducing short-term DNA double-strand breaks in the promoter regions of these genes. Topo IIβ is also involved in the release of paused RNA polymerase at specific sites, allowing for efficient progression to gene transcription. Together with other associated enzymes and proteins, topo IIβ ensures the controlled and transient up-regulation of gene expression in response to various signals.
Topoisomerase Inhibitors (Anti topoisomerase)
Topoisomerase inhibitors are a class of drugs that interfere with the activity of topoisomerases, thereby disrupting DNA replication and repair processes in cells. These inhibitors have been widely used in cancer treatment due to their ability to target rapidly dividing cancer cells. There are two main types of topoisomerase inhibitors: topoisomerase I inhibitors and topoisomerase II inhibitors.
- Topoisomerase I inhibitors: These drugs target and inhibit the activity of topoisomerase I enzymes, preventing them from relieving DNA supercoiling and causing DNA strand breaks. Examples of topoisomerase I inhibitors include:
- Camptothecins: Drugs such as irinotecan and topotecan are derived from the natural compound camptothecin. They stabilize the topoisomerase I-DNA complex, leading to the accumulation of DNA strand breaks and inhibition of DNA replication.
- Topoisomerase II inhibitors: These drugs interfere with the activity of topoisomerase II enzymes, which are responsible for DNA strand cleavage and re-ligation during DNA replication and repair. By blocking these enzymes, topoisomerase II inhibitors prevent DNA repair and cause DNA damage. Examples of topoisomerase II inhibitors include:
- Anthracyclines: Drugs such as doxorubicin and daunorubicin are widely used topoisomerase II inhibitors. They intercalate into DNA, preventing the re-ligation of DNA strands after topoisomerase II-mediated cleavage, leading to DNA breaks and inhibition of DNA replication.
- Etoposide: Etoposide is a topoisomerase II inhibitor that acts by binding to the topoisomerase II-DNA complex and stabilizing it in a cleaved state, resulting in DNA damage and inhibition of DNA replication.
Topoisomerase inhibitors are effective in killing cancer cells because cancer cells often exhibit higher rates of DNA replication and are more sensitive to DNA damage. By targeting topoisomerases, these drugs disrupt essential DNA processes in cancer cells, leading to cell death. However, it’s worth noting that these drugs can also affect normal, healthy cells, leading to side effects. Therefore, their use is carefully managed and monitored in clinical settings.
Topoisomerase Function – What is the function of topoisomerase?
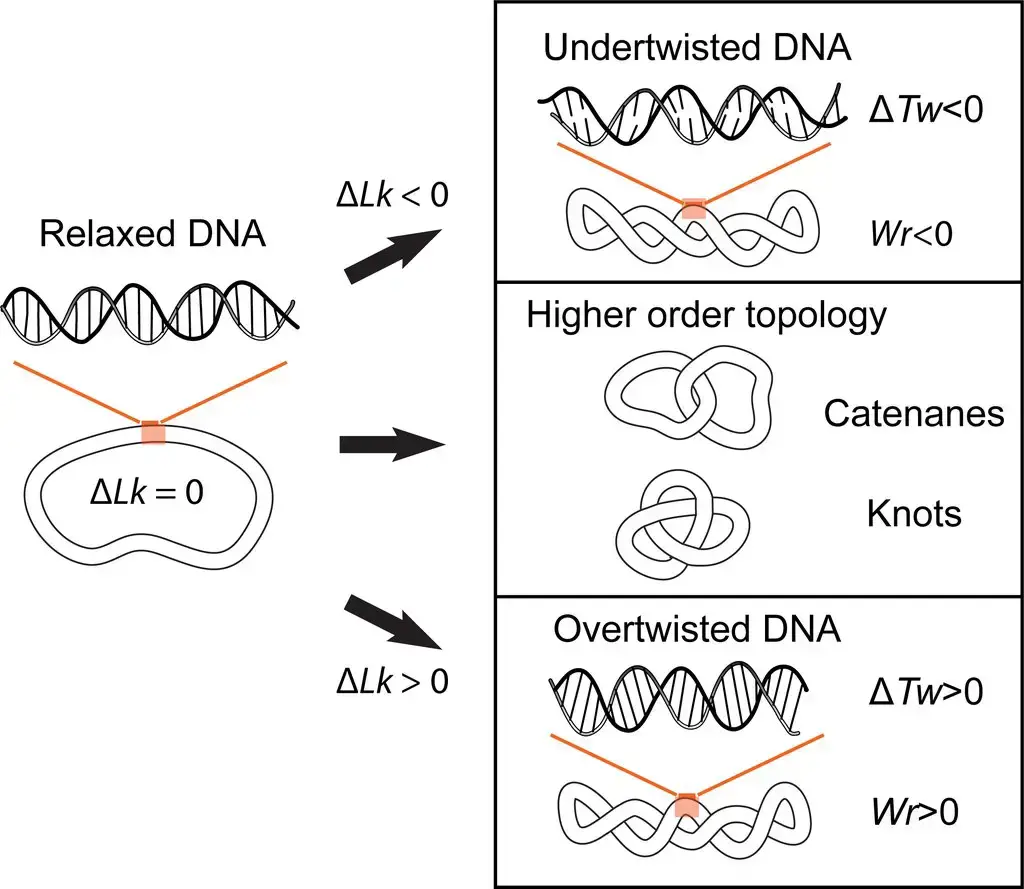
Topoisomerases are essential enzymes that play crucial roles in the maintenance and regulation of DNA structure and function. They are involved in various cellular processes that require changes in DNA topology, including DNA replication, transcription, recombination, and chromatin remodeling. The main functions of topoisomerases are as follows:
- DNA Unwinding and Relaxation: Topoisomerases are responsible for relieving torsional stress and supercoiling in DNA. During DNA replication and transcription, the unwinding of DNA strands can generate positive or negative supercoils ahead of the replication or transcription machinery. Topoisomerases can introduce transient breaks in the DNA strands to relax these supercoils, allowing DNA replication and transcription to proceed smoothly.
- Chromosome Condensation: Topoisomerases play a crucial role in condensing and organizing chromosomes during cell division. They help to remove excess supercoils and resolve DNA tangles and knots that can arise during DNA replication and recombination. By promoting proper chromosome condensation, topoisomerases ensure accurate chromosome segregation during cell division.
- DNA Segregation: Topoisomerases are involved in the segregation of replicated chromosomes during cell division. They help to resolve the intertwined DNA strands that arise when sister chromatids are replicated and entangled. By introducing transient DNA breaks and resealing them, topoisomerases enable the proper separation of replicated DNA strands and facilitate their segregation into daughter cells.
- DNA Repair and Recombination: Topoisomerases are critical for DNA repair and recombination processes. They can untangle and unlink DNA strands to facilitate the repair of DNA damage, such as DNA breaks or cross-links. Additionally, topoisomerases are involved in the resolution of DNA recombination intermediates and the exchange of genetic information between DNA molecules.
What does topoisomerase do?
Topoisomerases are enzymes that play a critical role in the maintenance and regulation of DNA structure and function. Their primary function is to manage and alter the topology of DNA molecules. DNA topology refers to the three-dimensional arrangement and organization of DNA strands, including the degree of supercoiling, knotting, and intertwining.
The main actions of topoisomerases can be summarized as follows:
- Supercoiling: Topoisomerases can relieve or induce supercoiling in DNA. Supercoiling occurs when the double helix of DNA is twisted upon itself, resulting in a more tightly or loosely wound structure. Topoisomerases can relax positive or negative supercoils by introducing transient breaks in the DNA strands and then resealing them.
- DNA Unwinding: During processes such as DNA replication, transcription, and DNA repair, DNA strands need to be unwound to provide access to the genetic information encoded within them. Topoisomerases help in the unwinding of DNA by creating breaks in one or both DNA strands, allowing the strands to rotate and unwind. This action prevents excessive DNA tangling and facilitates the progression of replication and transcription.
- DNA Segregation: When cells divide, replicated chromosomes need to be separated and segregated into daughter cells accurately. Topoisomerases play a crucial role in DNA segregation by resolving DNA entanglements and interlinkages that arise during DNA replication and recombination. They help in the separation and disentanglement of DNA strands, ensuring proper chromosome segregation.
- DNA Repair and Recombination: Topoisomerases are involved in DNA repair and recombination processes. They can untangle and unlink DNA strands to facilitate the repair of DNA damage, such as breaks or cross-links. Additionally, topoisomerases help in resolving DNA recombination intermediates, which are crucial for genetic recombination and the exchange of genetic information between DNA molecules.
Topoisomerase role in DNA replication
Topoisomerases play essential roles in DNA replication by resolving topological problems that arise during the process. DNA replication involves the unwinding of the double helix and the separation of the two DNA strands to allow for the synthesis of new complementary strands. However, this unwinding process can generate tension and supercoiling in the DNA molecule. Topoisomerases help to alleviate these topological stresses and ensure smooth progression of DNA replication.
Specifically, topoisomerases perform the following functions during DNA replication:
- Relaxation of Supercoils: As the DNA helix unwinds during replication, positive supercoiling can build up ahead of the replication fork, creating tension and hindering the unwinding process. Topoisomerases, particularly type I topoisomerases, act to relax these supercoils by introducing transient single-strand breaks in the DNA, allowing the strands to rotate and release the tension.
- Removal of DNA Knots and Tangles: During replication, the movement of the replication fork can generate DNA knots and tangles as the two parental strands separate. Topoisomerases, particularly type II topoisomerases, are involved in the decatenation of replicated DNA molecules by untangling and unlinking the intertwined DNA strands. They achieve this by introducing transient double-strand breaks and passing one DNA segment through another, resolving the knots and allowing proper separation of the replicated DNA molecules.
- Prevention of Replication Fork Collapse: Topoisomerases help to prevent replication fork collapse and stalling. If DNA replication encounters obstacles, such as DNA lesions or tightly bound proteins, the replication fork can stall or collapse, leading to DNA damage and genome instability. Topoisomerases, by relaxing supercoils and resolving DNA knots and tangles, help to alleviate the stress on the replication fork and prevent its collapse, ensuring the smooth continuation of DNA synthesis.
Overall, topoisomerases are critical players in DNA replication, ensuring the proper unwinding, relaxation, and separation of DNA strands to facilitate accurate and efficient replication of the genetic material.
Topoisomerase 1 vs 2
| Characteristic | Topoisomerase 1 | Topoisomerase 2 |
|---|---|---|
| Type | Type I | Type II |
| DNA Cleavage | Single-strand breaks | Double-strand breaks |
| ATP Requirement | Generally ATP-independent | ATP-dependent |
| Linking Number Change | Change by one unit | Change by two units |
| Substrate Specificity | Acts on single DNA strands | Acts on double-stranded DNA |
| Function in DNA Replication | Relaxes supercoils, resolves DNA knots, and prevents replication fork collapse | Relaxes supercoils, resolves DNA knots, and prevents replication fork collapse |
| Examples | Topoisomerase 1 (human), Topo IA (bacteria), Reverse Gyrase (archaea/bacteria) | Topoisomerase 2 (human), Topo II (eukaryotes), Topo IV (bacteria), DNA gyrase (bacteria) |
Topoisomerase vs Gyrase
Topoisomerase and gyrase are both enzymes involved in the regulation of DNA topology, but they have some differences in their characteristics and functions. Here is a comparison between topoisomerase and gyrase:
Topoisomerase:
- Topoisomerase is a broad class of enzymes that includes various types, such as topoisomerase I and topoisomerase II.
- It is a large class of enzymes involved in maintaining the topology of DNA by regulating the number of twists and supercoils in the DNA molecule.
- Topoisomerases can be found in both prokaryotes and eukaryotes, and they perform different functions in DNA replication, transcription, recombination, and chromosome condensation.
- They can introduce both negative and positive supercoiling in DNA, depending on the specific type and cellular context.
- Different types of topoisomerases may or may not be ATP-dependent, meaning they may or may not require ATP hydrolysis for their function.
Gyrase:
- Gyrase is a specific type of topoisomerase, belonging to the sub-class of Type II topoisomerases.
- It is primarily found in prokaryotes, such as bacteria, but can also be present in some eukaryotes.
- The main function of gyrase is to introduce negative supercoiling into DNA strands.
- Gyrase is an ATP-dependent enzyme, meaning it requires ATP hydrolysis for its activity.
- By introducing negative supercoils, gyrase helps in the compaction of DNA, regulates gene expression, and facilitates processes like DNA replication and transcription in prokaryotes.
In summary, gyrase is a type of topoisomerase that specifically introduces negative supercoiling in DNA strands, while topoisomerase is a broader class of enzymes that includes gyrase and other types. Topoisomerases can have various functions and may or may not be ATP-dependent.
| Characteristic | Topoisomerase | Gyrase |
|---|---|---|
| Classification | Broad class of enzymes | Type II topoisomerase |
| Types | Multiple types (e.g., topoisomerase I, topoisomerase II) | Subset of topoisomerase class |
| Presence | Found in both prokaryotes and eukaryotes | Mostly found in prokaryotes, some eukaryotes |
| Function | Maintains DNA topology | Introduces negative supercoiling in DNA |
| Supercoiling | Can introduce both negative and positive supercoiling | Specifically introduces negative supercoiling |
| ATP-dependence | Can be ATP-dependent or ATP-independent | ATP-dependent |
Topoisomerase vs Helicase
| Characteristic | Topoisomerase | Helicase |
|---|---|---|
| Function | Prevents supercoiling of DNA | Unwinds DNA strands |
| DNA vs. RNA | Acts on DNA only | Acts on DNA and RNA |
| Target of action | Phosphodiester bond in DNA backbone | Hydrogen bonds between DNA strands |
| Types | Type I Topoisomerase | RNA helicase |
| Type II Topoisomerase | DNA helicase |
FAQ
Where would the enzyme topoisomerase attach during dna replication?
During DNA replication, topoisomerase enzymes would attach to the DNA strands at specific sites to perform their functions. The exact attachment points and actions may vary depending on the type of topoisomerase involved (Type I or Type II). Here are the general locations where topoisomerases would attach during DNA replication:
Type I topoisomerases: These enzymes typically bind to the DNA at single-stranded regions or DNA loops. They can help relax supercoiled DNA, remove DNA knots and tangles, and prevent the collapse of replication forks. Type I topoisomerases, such as Topoisomerase 1 in humans, can interact with the DNA ahead of the replication fork to relieve torsional stress caused by the unwinding of the double helix.
Type II topoisomerases: These enzymes bind to the double-stranded DNA and are involved in the resolution of intertwined DNA molecules. During DNA replication, Type II topoisomerases, such as Topoisomerase 2 in humans, play a crucial role in managing DNA supercoiling and disentangling DNA strands. They are often found near the replication forks to resolve topological problems arising from DNA replication.
It’s important to note that the precise binding and action of topoisomerases during DNA replication can be complex and can vary depending on the specific cellular context and the organism being studied. Additionally, different types and subtypes of topoisomerases may have distinct roles and specific binding sites.
Which type of topoisomerase requires the energy of atp hydrolysis for function? why?
Type II topoisomerases require the energy of ATP hydrolysis for their function. This is because the mechanism of action of Type II topoisomerases involves breaking both strands of DNA simultaneously, passing another DNA duplex through the gap, and then resealing the DNA strands. These processes require a significant amount of energy to overcome the resistance of the DNA double helix and facilitate the strand passage.
ATP hydrolysis provides the necessary energy to drive the conformational changes in Type II topoisomerases. The ATP molecule is hydrolyzed to ADP and inorganic phosphate (Pi), releasing energy that can be utilized by the enzyme to perform its functions. The energy from ATP hydrolysis is used to induce the conformational changes required for DNA cleavage, strand passage, and DNA resealing.
In addition to providing energy, ATP hydrolysis also regulates the activity of Type II topoisomerases. The binding and hydrolysis of ATP help coordinate the enzymatic cycle, ensuring the proper timing and sequence of events during DNA manipulation.
In summary, the energy from ATP hydrolysis is necessary for the function of Type II topoisomerases as it enables the breaking and resealing of DNA strands and facilitates the passage of DNA through the enzyme.
What does topoisomerase 2 do?
Topoisomerase 2, also known as DNA topoisomerase II, is an enzyme that plays a crucial role in DNA metabolism. Here are some key functions of topoisomerase 2:
DNA Unwinding: Topoisomerase 2 is involved in the unwinding of DNA during processes like DNA replication and transcription. It helps relieve the torsional stress that builds up ahead of the replication fork or RNA polymerase by introducing temporary double-stranded breaks in the DNA.
DNA Decatenation: Topoisomerase 2 is responsible for decatenating DNA, which refers to the separation of interlinked or intertwined DNA molecules. During DNA replication, sister chromatids are initially intertwined, and topoisomerase 2 resolves these interlinks to separate the replicated DNA molecules.
Chromosome Condensation: Topoisomerase 2 plays a crucial role in the condensation of chromosomes during cell division. It helps to compact and organize DNA into the characteristic condensed structures seen during mitosis and meiosis.
Regulation of DNA Topology: Topoisomerase 2 is involved in maintaining the proper topology of DNA by altering the number of twists and supercoils in the DNA molecule. It can introduce both negative and positive supercoils into the DNA, depending on the specific cellular context and requirements.
Repair of DNA Damage: Topoisomerase 2 participates in DNA repair processes by resolving DNA knots and tangles that can occur due to DNA damage, such as DNA double-strand breaks. It helps in the proper rejoining of DNA strands and the restoration of the DNA structure.
Overall, topoisomerase 2 plays essential roles in DNA replication, chromosome dynamics, and DNA repair, ensuring the proper functioning and integrity of the genome.
Where would the enzyme topoisomerase attach during dna replication?
During DNA replication, topoisomerases, including DNA topoisomerase I and DNA topoisomerase II, play important roles in resolving the topological constraints that arise as a result of DNA unwinding and replication. These enzymes help relieve torsional strain and prevent DNA entanglements during the replication process.
DNA topoisomerase I acts by creating transient single-strand breaks in the DNA molecule. It cleaves one strand of the DNA double helix, allowing the other strand to rotate around the intact strand, thereby releasing the torsional stress. Once the rotation is complete, the enzyme reseals the DNA strand break, and the replication process can continue.
DNA topoisomerase II, on the other hand, is involved in managing the entanglements that occur when the DNA molecule is completely unwound during replication. It introduces transient double-strand breaks in the DNA helix and passes one segment of DNA through another, effectively untangling the DNA strands. Once the entanglement is resolved, the enzyme reseals the double-strand breaks, ensuring the integrity of the DNA molecule.
Therefore, DNA topoisomerases, including topoisomerase I and topoisomerase II, attach and act on the DNA molecule during replication to alleviate the torsional stress and entanglements that can arise during the unwinding and copying of the DNA strands.
References
- Goedecke, W. (2007). Topoisomerases. xPharm: The Comprehensive Pharmacology Reference, 1–2. doi:10.1016/b978-008055232-3.60609-9
- Majumder, H. K. (2013). Topoisomerases. Brenner’s Encyclopedia of Genetics, 78–79. doi:10.1016/b978-0-12-374984-0.01545-x
- Hwang, J., & Hwong, C.-L. (1994). Cellular Regulation of Mammalian DNA Topoisomerases. Advances in Pharmacology, 167–189. doi:10.1016/s1054-3589(08)60545-1
- Drolet, M., Wu, H.-Y., & Liu, L. F. (1994). Roles of DNA Topoisomerases in Transcription. Advances in Pharmacology, 135–146. doi:10.1016/s1054-3589(08)60543-8
- RANDONE, S. B., GUIDUCCI, S., & CERINIC, M. M. (2007). TOPOISOMERASE-I (SCL-70) AUTOANTIBODIES. Autoantibodies, 231–238. doi:10.1016/b978-044452763-9/50035-4