What is TaqMan Probe?
- TaqMan probes are a sophisticated tool used to enhance the specificity of quantitative Polymerase Chain Reaction (qPCR) assays. This method, originally introduced by Kary Mullis at Cetus Corporation in 1991, has been further refined for both diagnostic and research purposes by companies such as Hoffmann-La Roche and Applied Biosystems (now part of Thermo Fisher Scientific).
- The TaqMan probe mechanism leverages the 5´–3´ exonuclease activity of Taq polymerase. This enzyme activity is crucial for the probe’s function, as it cleaves a dual-labeled probe during its hybridization to a complementary target sequence. The probe is labeled with a reporter dye at one end and a quencher dye at the other. When the probe is intact, the proximity of the quencher dye suppresses the fluorescence of the reporter dye. During the PCR process, as Taq polymerase extends the primer, it encounters the TaqMan probe bound to the target sequence. The polymerase’s exonuclease activity cleaves the probe, separating the reporter dye from the quencher dye and thus allowing the reporter dye to emit fluorescence. This fluorescence can then be measured, providing a real-time indication of the quantity of the target DNA.
- The primary advantage of using TaqMan probes in qPCR is the increased specificity of detection. Unlike other qPCR methods, the TaqMan approach reduces the likelihood of non-specific amplification signals. This specificity is particularly important in clinical diagnostics and research settings where precise quantification of DNA or RNA is critical.
- Interestingly, the name “TaqMan” is derived from a play on the video game Pac-Man, combined with Taq polymerase. This name reflects the probe’s mechanism, which is reminiscent of the Pac-Man character’s method of eating pellets, analogous to Taq polymerase’s process of cleaving the probe during PCR amplification.
- In summary, TaqMan probes represent a significant advancement in the field of molecular biology, providing a reliable and specific method for quantifying nucleic acids. Their development has greatly enhanced the precision of quantitative PCR, making them a valuable tool in both research and diagnostic applications.
Definition of TaqMan
TaqMan probes are hydrolysis probes used in quantitative PCR (qPCR) to increase specificity by utilizing the 5´–3´ exonuclease activity of Taq polymerase to cleave a dual-labeled probe during hybridization, resulting in a fluorescence signal that allows for precise quantification of the target DNA.
Principle of TaqMan
TaqMan technology is a highly specific method used in quantitative PCR (qPCR) to detect and quantify nucleic acids. The principle behind TaqMan probes involves the use of fluorescently labeled oligonucleotide probes that enhance the accuracy of DNA amplification measurements. Here is a detailed explanation of the TaqMan principle:
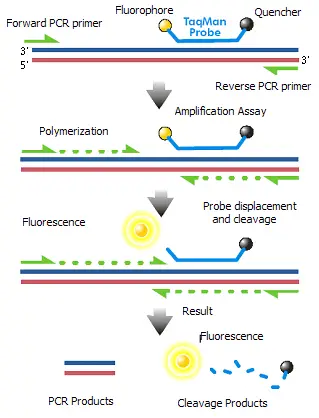
- Probe Structure:
- Fluorophore: Attached to the 5’-end of the oligonucleotide probe. Common fluorophores include 6-carboxyfluorescein (FAM) and tetrachlorofluorescein (TET).
- Quencher: Attached to the 3’-end of the probe. Quenchers like tetramethylrhodamine (TAMRA) absorb the fluorescence emitted by the fluorophore through a process called Förster resonance energy transfer (FRET) when they are in close proximity.
- Mechanism:
- Binding: TaqMan probes are designed to anneal specifically within the DNA region that is being amplified by a set of primers. This binding occurs on single-stranded DNA.
- Minor Groove Binder (MGB): Some TaqMan probes are conjugated with a minor groove binder, such as dihydrocyclopyrroloindole tripeptide (DPI3). The MGB increases the binding affinity of the probe to the target sequence, raising the melting temperature (Tm) and enhancing probe stability through van der Waals forces.
- PCR Process:
- Extension and Cleavage: During the PCR, as Taq polymerase extends the primer to synthesize the new DNA strand, its 5′ to 3′ exonuclease activity degrades the bound TaqMan probe.
- Fluorescence Release: The degradation of the probe separates the fluorophore from the quencher, relieving the quenching effect. This separation allows the fluorophore to emit fluorescence when excited by the thermal cycler’s light source.
- Detection: The emitted fluorescence is directly proportional to the amount of probe degradation, which correlates with the quantity of the target DNA template in the PCR reaction.
- Fluorescence Measurement:
- The increase in fluorescence is measured at each cycle of the PCR, providing a real-time indication of DNA amplification. The amount of fluorescence detected is directly proportional to the amount of target DNA present, allowing for precise quantification.
Applications of TaqMan
- Gene Expression Assays:
- TaqMan probes are used to quantify gene expression levels by measuring the amount of mRNA present in a sample. This helps in understanding gene regulation and function in various biological processes.
- Pharmacogenomics:
- In pharmacogenomics, TaqMan assays help identify genetic variations that influence individual responses to drugs. This information can be used to tailor medical treatments to achieve better therapeutic outcomes.
- Human Leukocyte Antigen (HLA) Genotyping:
- TaqMan assays are employed in HLA genotyping to determine compatibility for organ transplantation. Accurate HLA typing is crucial for matching donors and recipients to minimize transplant rejection.
- Determination of Viral Load in Clinical Specimens:
- TaqMan probes are used to quantify the amount of viral RNA or DNA in clinical samples, such as those from patients with HIV or Hepatitis. Monitoring viral load is essential for diagnosing infections and evaluating the effectiveness of antiviral therapies.
- Bacterial Identification:
- TaqMan assays can identify bacterial species in clinical or environmental samples. This application is important for diagnosing bacterial infections and conducting microbial research.
- DNA Quantification:
- TaqMan probe-based qPCR is a reliable method for measuring the amount of DNA in a sample. This is useful in various fields, including forensic science, where precise DNA quantification is necessary for genetic analysis.
- SNP Genotyping:
- Single nucleotide polymorphisms (SNPs) are variations at a single position in a DNA sequence among individuals. TaqMan assays can detect and genotype these SNPs, which is valuable in genetic studies, including those related to disease susceptibility and population genetics.
- Verification of Microarray Results:
- Microarrays are used to analyze gene expression or genetic variations across many genes simultaneously. TaqMan qPCR is often used to verify and validate the results obtained from microarray experiments, ensuring accuracy and reliability.
Advantages of TaqMan
- High Specificity: TaqMan probes are designed to bind specifically to the target sequence, minimizing non-specific amplification and improving the accuracy of quantification.
- Sensitivity: TaqMan assays can detect low levels of target DNA or RNA, making them suitable for applications where high sensitivity is required, such as detecting viral load in clinical specimens.
- Quantitative Accuracy: The real-time monitoring of fluorescence during PCR allows for precise quantification of the target nucleic acid, providing quantitative data that can be compared across samples.
- Speed and Efficiency: TaqMan assays are rapid and can be completed in a relatively short time, allowing for high-throughput analysis of multiple samples.
- Automation Compatibility: TaqMan assays can be easily automated, reducing the risk of human error and increasing throughput in laboratories with high sample volumes.
- Versatility: TaqMan technology can be used for various applications, including gene expression analysis, SNP genotyping, viral load determination, and microbiological identification.
- Cost-Effectiveness: While initial setup costs for TaqMan assays may be higher compared to other PCR methods, the high specificity and sensitivity can reduce the need for repeated testing, making it a cost-effective choice in the long run.
- Compatibility with Multiplexing: TaqMan assays can be multiplexed, allowing for the simultaneous detection and quantification of multiple targets in a single reaction, saving time and resources.
- Standardization: TaqMan assays are well-established and standardized, with protocols and reagents readily available from various manufacturers, ensuring consistent and reliable results across different laboratories.
References
- The Real-Time TaqMan PCR and Applications in Veterinary Medicine – From PacMan to TaqMan – a computer game revisited Archived 2009-05-05 at the Wayback Machine
- Holland, P. M.; Abramson, R. D.; Watson, R.; Gelfand, D. H. (1991). “Detection of specific polymerase chain reaction product by utilizing the 5′—-3′ exonuclease activity of Thermus aquaticus DNA polymerase”. Proceedings of the National Academy of Sciences of the United States of America. 88 (16): 7276–7280. Bibcode:1991PNAS…88.7276H. doi:10.1073/pnas.88.16.7276. PMC 52277. PMID 1871133.
- TaqMan Gene Expression – NCBI Projects
- TaqMan Probes: Introduction, functioning and applications
- Bustin SA (October 2000). “Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays”. J. Mol. Endocrinol. 25 (2): 169–93. doi:10.1677/jme.0.0250169. PMID 11013345.
- Kutyavin IV, Afonina IA, Mills A, Gorn VV, Lukhtanov EA, Belousov ES, Singer MJ, Walburger DK, Lokhov SG, Gall AA, Dempcy R, Reed MW, Meyer RB, Hedgpeth J (2000). “3′-Minor groove binder-DNA probes increase sequence specificity at PCR extension temperatures”. Nucleic Acids Res. 28 (2): 655–661. doi:10.1093/nar/28.2.655. PMC 102528. PMID 10606668.
- AlleleID – Assay Design for Bacterial Identification
- https://www.ncbi.nlm.nih.gov/probe/docs/projtaqman/