Coronaviruses are a type of virus that can cause illness, ranging from the common cold to more severe diseases such as Middle East Respiratory Syndrome (MERS) and Severe Acute Respiratory Syndrome (SARS). Coronaviruses are so named because they have a crown-like appearance when viewed under a microscope. They are primarily transmitted from person to person through respiratory droplets, such as when an infected person coughs or sneezes. Symptoms of coronavirus infection can include fever, cough, and difficulty breathing. Some strains of coronavirus can be severe and even fatal, particularly for older adults or people with underlying health conditions. There is currently no specific treatment for coronavirus infection, but symptoms can be managed with supportive care. It is important to practice good hygiene, such as washing your hands frequently and covering your mouth and nose when you cough or sneeze, to help prevent the spread of coronavirus.
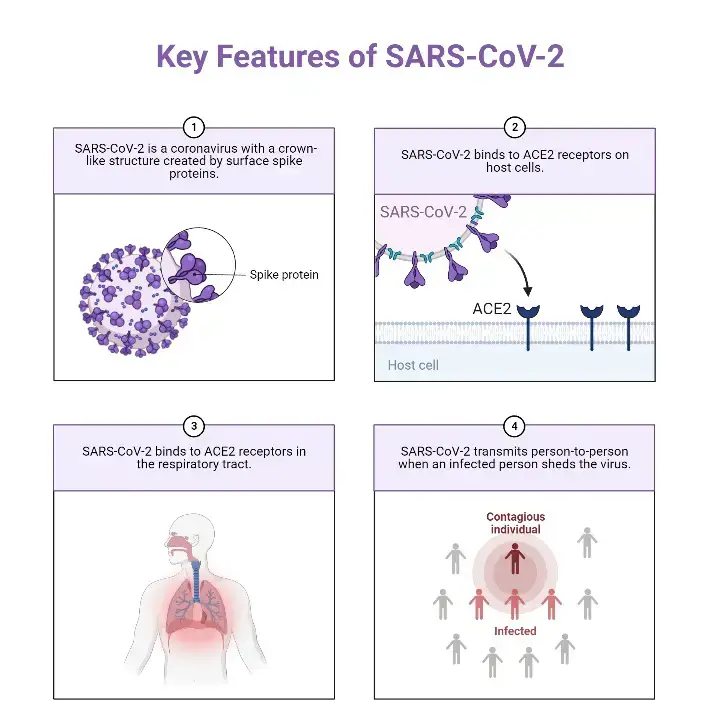
Characteristics of SARS-CoV-2 (COVID-19)
- SARS-CoV-2 is the virus that causes the disease COVID-19. The virus is an enveloped virus, which means it is surrounded by a protective outer layer called an envelope. The envelope is made up of a lipid bilayer, which is composed of two layers of fat molecules.
- Inside the envelope, the virus has a capsid, which is a protein shell that encloses the viral genome. The capsid is made up of multiple copies of a single protein called the spike protein. The spike protein is what gives the virus its crown-like appearance and allows it to bind to and enter host cells.
- The genome of SARS-CoV-2 is a single-stranded RNA molecule. It contains the instructions for synthesizing all of the proteins that the virus needs to replicate and infect cells.
- SARS-CoV-2 is a relatively large virus, measuring about 80-120 nanometers in diameter. It is classified as a betacoronavirus, which is a subfamily of coronaviruses that also includes MERS-CoV and SARS-CoV.
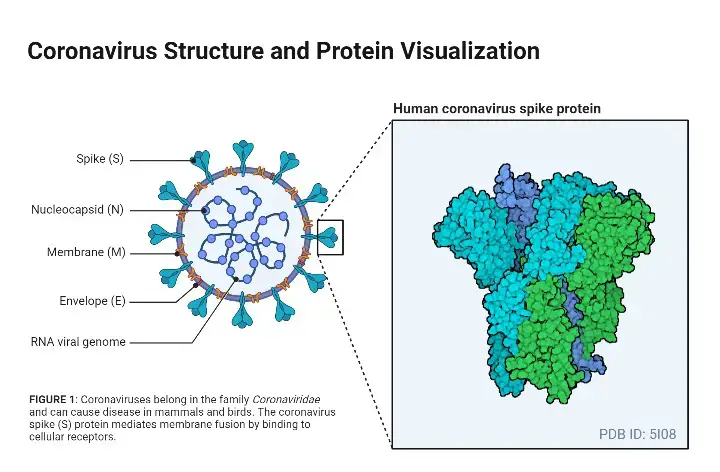
Structure of SARS-CoV-2
- Each SARS-CoV-2 virion has a diameter of 60–140 nanometers (2.4106–5.5109 in); Its estimated mass within the worldwide human population ranges between 0.1 and 10 kg.
- In the family Coronaviridae, the subfamily Coronavirinae has four genera: Alphacoronavirus, Betacoronavirus, Gammacoronavirus, and Deltacoronavirus.
- The genome of CoVs (27–32 kb) is a single-stranded positive-sense RNA (+ssRNA) that is larger than the genomes of any other RNA viruses.
- The nucleocapsid protein (N) forms the capsid outside of the genome, and the genome is further encapsulated by an envelope composed of three structural proteins: membrane protein (M), spike protein (S), and envelope protein (E).
- As a member of the coronavirus family, the recently sequenced SARS-CoV-2 genome measures roughly 29.9 kb.
- Four structural proteins (S, E, M, and N) and sixteen non-structural proteins (nsp116) are present in SARS-CoV-2. Nsp1 mediates RNA replication and processing.
- Nsp2 controls the host cell’s survival signalling pathway. It is thought that Nsp3 separates the translated protein. Nsp4 contains transmembrane domain 2 (TM2) and changes ER membranes. Nsp5 is involved in the polyprotein pathway during replication.
- Nsp6 is a putative transmembrane domain. The presence of nsp7 and nsp8 substantially enhanced the association between nsp12 and template-primer RNA.
- Nsp9 serves as a protein that binds ssRNA. Nsp10 is essential for viral mRNA cap methylation.
- Nsp12 contains RNA-dependent RNA polymerase (RdRp), an essential component of coronavirus replication/transcription.
- Nsp13 interacts with ATP, and its zinc-binding domain is involved in the replication and transcription processes.
- Nsp14 is an exoribonuclease with a proofreading domain. Nsp15 possesses endoribonuclease activity depending on Mn(2+). 2′-O-ribose methyltransferase is Nsp16.
- Inhibition of host defences by NSP-mediated effects on splicing, translation, and protein trafficking is demonstrated in one study.
- NSP16 binds the mRNA recognition domains of the U1 and U2 snRNAs to inhibit mRNA splicing in response to SARS-CoV-2 infection. NSP1 binds to 18S ribosomal RNA in the ribosome’s mRNA entry channel in order to inhibit mRNA translation.
- NSP8 and NSP9 bind to 7SL RNA at the Signal Recognition Particle to impede protein trafficking to the cell membrane. Based on their architectures, the following SARS-CoV-2 proteins could possibly serve as antiviral therapeutic targets.
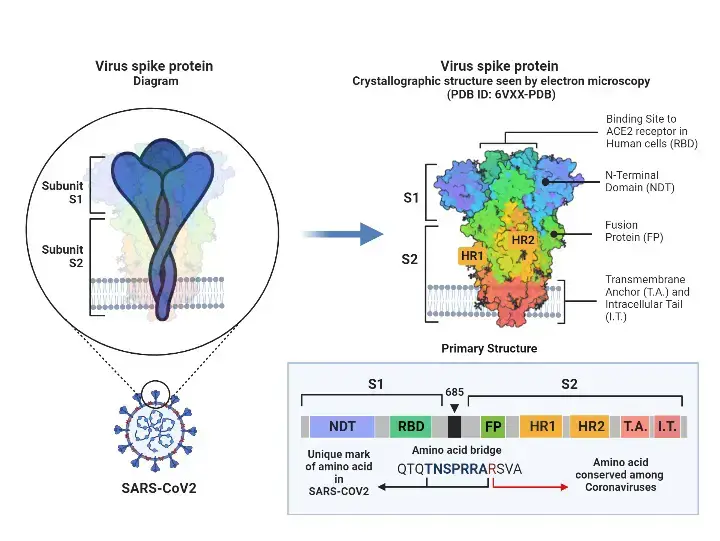
Spike Glycoprotein
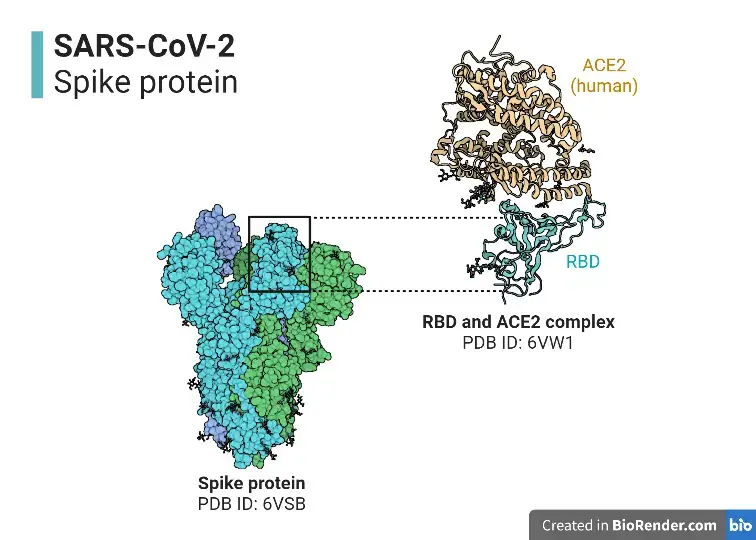
- S protein (spike glycoprotein) facilitates the entry of coronaviruses into host cells. Homotrimeric transmembrane spike glycoproteins protrude from the viral surface.
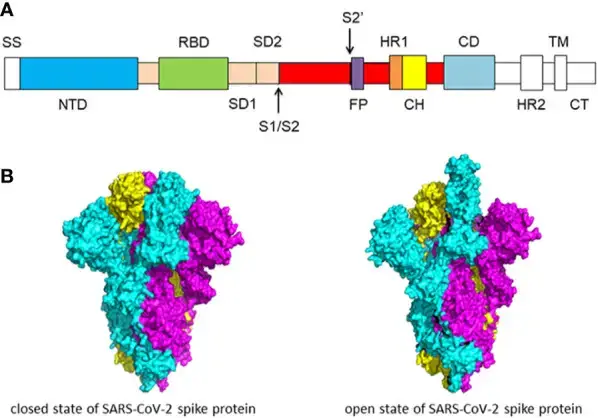
- The spike glycoprotein is essential for the entrance of coronaviruses, making it a desirable target for antiviral agents. The S protein consists of two functional subunits, S1 and S2.
- The N-terminal domain (NTD) and receptor binding domain make up the S1 subunit (RBD). S1 subunit’s job is to attach to the receptor on the host cell.
- Fusion peptide (FP), heptad repeat 1 (HR1), central helix (CH), connector domain (CD), heptad repeat 2 (HR2), transmembrane domain (TM), and cytoplasmic tail (CT) are components of the S2 subunit.
- The S2 component is responsible for fusing the membranes of viruses and host cells. S1/S2 protease cleavage site refers to the cleavage location at the interface between S1 and S2 subunits.
- Host proteases cut the spike glycoprotein at the S2′ cleavage site for all coronaviruses in order to activate the proteins, which are essential for fusing the membranes of viruses and host cells through irreversible conformational changes.
- N-linked glycans are essential for correct folding, antibody neutralisation, and extensively coating the spike protein trimers.
- SARS-CoV-2 S protein is structurally similar to the closely related SARS-CoV S protein. S1 and S2 subunits are non-covalently coupled in the prefusion state. Different types of coronaviruses utilise unique domains in the S1 subunit to identify distinct entrance receptors.
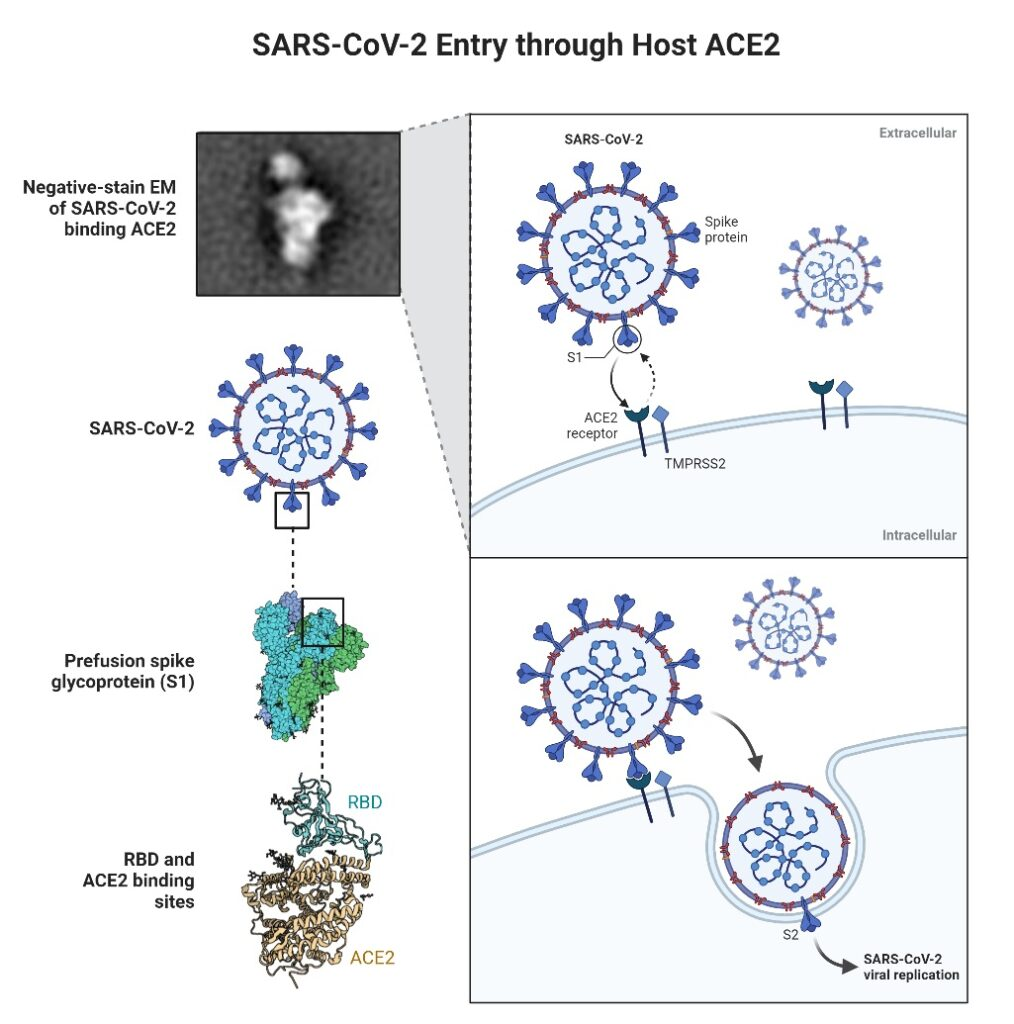
- In order to enter host cells, SARS-CoV and SARS-CoV-2 recognise the receptor angiotensin-converting enzyme 2 (ACE2) via the receptor binding domain (RBD).
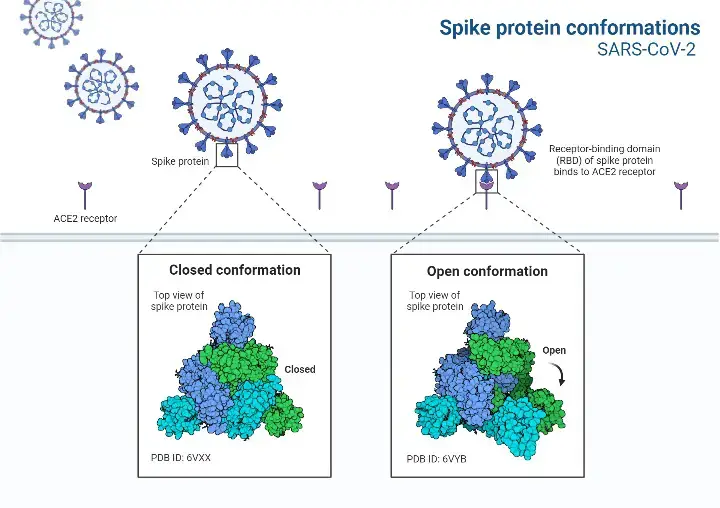
- The S protein has two structural configurations, including the closed and open states. The three recognition motifs do not protrude from the interface created by three spike protein protomers in the closed state.
- The RBD is in the “up” conformation when it is in the open position. The open state is required for the fusion of the SARS-CoV-2 and host cell membranes, allowing the virus to enter the host cells .
HR1 and HR2
- The 6-HB is produced by HR1 and HR2 and is essential for membrane fusion, which is dominated by the spike protein of SARS-CoV or SARS-CoV-2, making HR1 and HR2 a viable therapeutic target.
- The difference between the 6-HB of SARS-CoV-2 and SARS-CoV may solidify the 6-HB conformation of SARS-CoV-2 and strengthen the interactions between HR1 and HR2, hence increasing the infectiousness of SARS-CoV-2.
- The HR1-L6-HR2 complex comprises the majority of the HR1 and HR2 domains together with a linker. This fusion protein has a rod-like structure and is the usual 6-HB structure.
- Three HR1 domains come together in parallel to form a spiral coil trimer. The antiparallel intertwining of three HR2 domains around the coiled-coil core is mostly mediated by hydrophobic force. The hydrophobic residues on the HR2 dome attach to the hydrophobic groove created by each pair of adjacent HR1 helices.
- The S2 subunit of SARS-CoV and SARS-CoV-2 share an extremely identical 6-HB structure. HR1 of SARS-CoV and SARS-CoV-2 is 96% same, while HR2 is 100% identical.
- In the fusion core area of HR1 domain, there are eight unique residues. In the HR1 domain of SARS-CoV, lysine 911 forms a salt bridge with glutamic acid 1176 in the HR2 domain.
- Regarding SARS-CoV-2, a strong hydrogen bond replaces the salt bridge between serine 929 in HR1 and serine 1,196 in HR2.
- HR1 glutamine 915 has no interaction with HR2 in SARS-CoV However, there is a salt bridge between lysine 933 in HR1 and asparagine 1,192 in HR2 in SARS-CoV-2.
- There is a weak salt bridge between glutamic acid 918 in HR1 and arginine 1,166 in the SARS-CoV. Through a salt bridge, aspartic acid 936 in the HR1 of SARS-CoV-2 binds to arginine 1158.
- In the HR2 domain of the SARS-CoV, lysine 929 binds to glutamic acid 1,163 through a salt bridge, whereas threonine 925 does not bind to glutamic acid 1,163.
- However, the SARS-CoV-2 serine 943 and lysine 947 attach to glutamic acid 1,182 in HR2 via a hydrogen bond and a salt bridge. These changes may boost the infectiousness of SARS-CoV-2.
The Receptor Binding Domain (RBD)
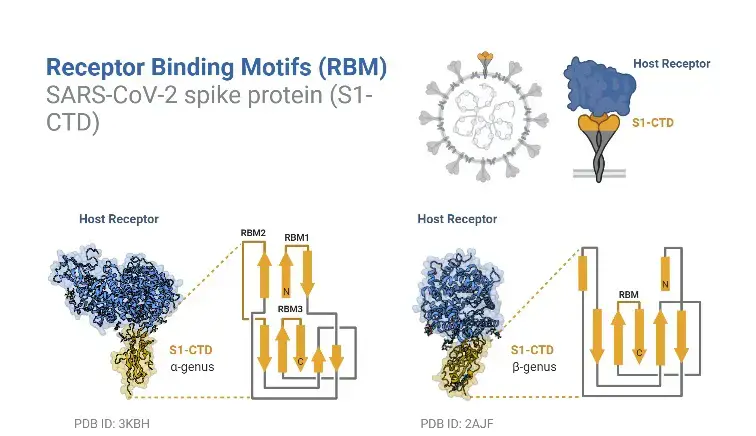
- The spike protein of SARS-CoV-2 has an RBD that particularly binds to the receptor ACE2. RBD is an important target for antiviral agents and antibodies. Two structural domains comprise the RBD of SARS-CoV-2: the core and the exterior subdomains.
- Core subdomain has a high level of conservation. It is composed of five antiparallel beta strands and a disulfide connection between two beta strands. The loop which is stabilised by a disulfide bond predominates the exterior subdomain.
- The SARS-CoV-2 RBD core is composed of five antiparallel beta sheets joined by loops and short helices. Between the antiparallel β4 and β7 strands is a receptor-binding motif (RBM) made up of loops and helices, as well as short β5 and β6 strands. RBM includes the majority of SARS-CoV-2 and ACE2 binding sites.
- In the RBD, eight of the nine Cys residues form four pairs of disulfide bonds. The presence of three disulfide bonds in the core of RBD increases the stability of the β sheet (C336-C361, C379-C432, and C391-C525).
- Regarding the remaining disulfide bond (C480-C488), it supports the inter-loop connections in RBM. The N-terminal peptidase domain of ACE2 comprises the binding site, which is formed by two RBM and ACE2 lobes.
- RBM binds to the tiny lobe on the underside of the ACE2 protein. RBM’s surface is somewhat curved inward to accommodate ACE2.
RBD-ACE2 Complex
- It is essential to comprehend the SARS-CoV-2 receptor recognition process, which dictates the virus’ infectiousness, host range, and pathogenesis. SARS-CoV-2 and SARS-CoV both identify ACE2 in humans.
- It has been discovered the crystal structure of SARS-CoV-2 RBD linked to ACE2. The overall method of combination of SARS-CoV-2 RBD-ACE2 complex is extremely similar to that of the previously identified SARS-CoV RBD-ACE2 complex.
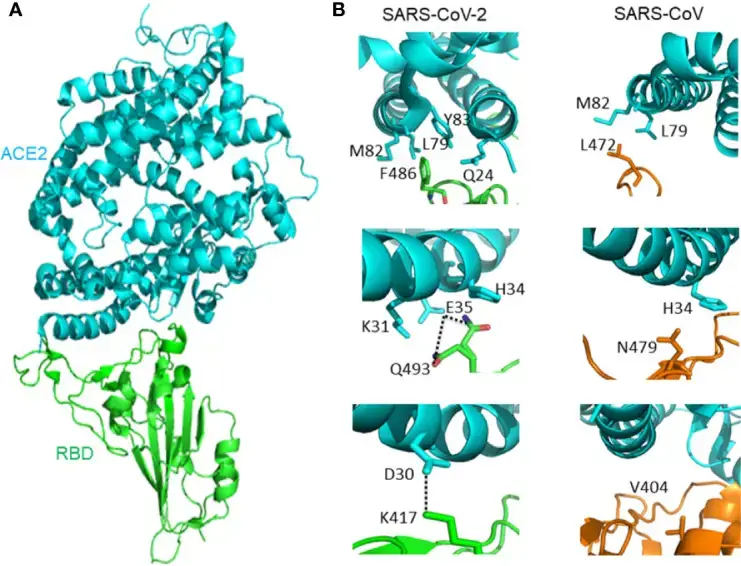
- Seventeen out of the twenty ACE2 residues that interact with the RBD of SARS-CoV and SARS-CoV-2 are identical. The binding affinity of SARS-CoV-2 and SARS-CoV RBD to ACE2 differs, however, due to minor interactions between ACE2 molecules.
- Greater affinity exists between ACE2 and SARS-CoV-2 than between ACE2 and SARS-CoV. SARS-CoV-2 F486 interacts with ACE2 Q24, L79, M82, and Y83 at the F486/L472 location, whereas SARS-CoV-2 L472 only interacts with ACE2 L79 and M82.
- SARS-CoV-2 Q493 interacts with ACE2 K31, E35, and H34 at location Q493/N479. Hydrogen bonds exist between Q493 and E35.
- SARS-CoV N479 interacts only with ACE2 H34. ACE2 D30 and SARS-CoV-2 K417 are connected by a salt bridge outside SARS-CoV-2 RBM. The SARS-CoV V404 did not contribute in ACE2 binding, however.
Furin Cleavage Site of the Spike Protein
- The S1/S2 boundary of SARS-CoV-2 spike protein is the region of cleavage by the subtilisin-like host cell protease furin, which distinguishes SARS-CoV-2 S from SARS-CoV S.
- The furin cleavage site consists of four residues (P681, R684, R683, and A684) and is positioned at the S1-S2 subunit interface. R682, R683, A684, and R685 serve as the minimal polybasic furin cleavage site, RXYR, where X or Y is a positively charged arginine or lysine.
- Such polybasic cleavage sites are absent in SARS-CoV and SARS-CoV-related group 2b betacoronaviruses found in humans, which may contribute to the high virulence of SARS-CoV-2 due to the fact that the furin proteases required for proteolytic activation of S are ubiquitously expressed in humans, resulting in expanded tissue tropism and pathogenesis.
- A study has also produced a SARS-CoV-2 mutant virus lacking the furin cleavage site (δPRRA) in the spike protein. In Vero E6 cells, the mutant virus processed spike proteins less efficiently than the wild-type SARS-CoV-2 virus.
- Additionally, the mutated virus replicated less efficiently in Calu3 human respiratory cells and caused less severe sickness in a hamster pathogenesis model. These results demonstrated the significance of the furin cleavage site in the replication and pathogenesis of SARS-CoV-2.
Genomic Organization of SARS-CoV-2
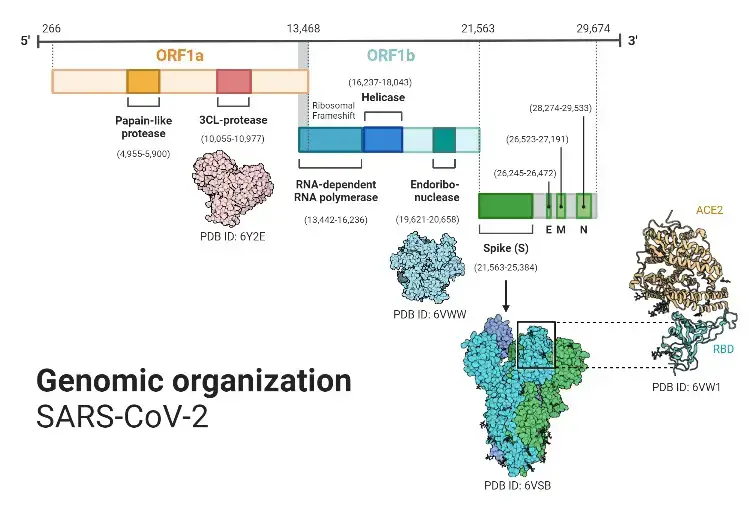
- Approximately 7 million SARS-CoV-2 genomes had been sequenced and deposited in public databases by the beginning of 2022, and another 800,000 or more were contributed monthly.
- SARS-CoV-2 contains a 30,000-base-long linear, positive-sense, single-stranded RNA genome. As with other coronaviruses, its genome is biassed against cytosine (C) and guanine (G) nucleotides. The genome is composed primarily of U (32.2%), followed by A (29.9%), G (19.6%), and C (18.9%) in similar proportions.
- The mutation of guanines and cytosines into adenosines and uracils, respectively, causes the nucleotide bias.
- It is believed that the alteration of CG dinucleotides occurs to evade the zinc finger antiviral protein associated cellular defence mechanism and to reduce the energy required to unbind the genome during replication and translation (adenosine and uracil base pair via two hydrogen bonds, cytosine and guanine via three).
- Due to the reduction of CG dinucleotides in its genome, the virus exhibits a significant codon use bias. For instance, the relative synonymous codon usage of arginine’s six distinct codons is AGA (2.67), CGU (1.46), AGG (.81), CGC (.58), CGA (.29), and CGG (.19). Similar codon use bias is observed in other coronaviruses connected to SARS.
References
- V’kovski P, Kratzel A, Steiner S, Stalder H, Thiel V (March 2021). “Coronavirus biology and replication: implications for SARS-CoV-2”. Nature Reviews. Microbiology. 19 (3): 155–170. doi:10.1038/s41579-020-00468-6. PMC 7592455. PMID 33116300.
- Zhu N, Zhang D, Wang W, Li X, Yang B, Song J, et al. (February 2020). “A Novel Coronavirus from Patients with Pneumonia in China, 2019”. The New England Journal of Medicine. 382 (8): 727–733. doi:10.1056/NEJMoa2001017. PMC 7092803. PMID 31978945.
- “Phylogeny of SARS-like betacoronaviruses”. nextstrain. Archived from the original on 20 January 2020. Retrieved 18 January 2020.
- Wong AC, Li X, Lau SK, Woo PC (February 2019). “Global Epidemiology of Bat Coronaviruses”. Viruses. 11 (2): 174. doi:10.3390/v11020174. PMC 6409556. PMID 30791586.
- Singh D, Yi SV (April 2021). “On the origin and evolution of SARS-CoV-2”. Experimental & Molecular Medicine. 53 (4): 537–547. doi:10.1038/s12276-021-00604-z. PMC 8050477. PMID 33864026.
- Jackson B, Boni MF, Bull MJ, Colleran A, Colquhoun RM, Darby AC, et al. (September 2021). “Generation and transmission of interlineage recombinants in the SARS-CoV-2 pandemic”. Cell. 184 (20): 5179–5188.e8. doi:10.1016/j.cell.2021.08.014. PMC 8367733. PMID 34499854. S2CID 237099659.
- “CoV2020”. GISAID EpifluDB. Archived from the original on 12 January 2020. Retrieved 12 January 2020.
- Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H (May 2020). “The Architecture of SARS-CoV-2 Transcriptome”. Cell. 181 (4): 914–921.e10. doi:10.1016/j.cell.2020.04.011. PMC 7179501. PMID 32330414.
- Hossain, Md. Golzar; Tang, Yan‐dong; Akter, Sharmin; Zheng, Chunfu (May 2022). “Roles of the polybasic furin cleavage site of spike protein in SARS‐CoV‐2 replication, pathogenesis, and host immune responses and vaccination”. Journal of Medical Virology. 94 (5): 1815–1820. doi:10.1002/jmv.27539. PMID 34936124. S2CID 245430230.
- To KK, Sridhar S, Chiu KH, Hung DL, Li X, Hung IF, et al. (December 2021). “Lessons learned 1 year after SARS-CoV-2 emergence leading to COVID-19 pandemic”. Emerging Microbes & Infections. 10 (1): 507–535. doi:10.1080/22221751.2021.1898291. PMC 8006950. PMID 33666147.
- Jackson CB, Farzan M, Chen B, Choe H (January 2022). “Mechanisms of SARS-CoV-2 entry into cells”. Nature Reviews Molecular Cell Biology. 23 (1): 3–20. doi:10.1038/s41580-021-00418-x. PMC 8491763. PMID 34611326.
- Braun E, Sauter D (2019). “Furin-mediated protein processing in infectious diseases and cancer”. Clinical & Translational Immunology. 8 (8): e1073. doi:10.1002/cti2.1073. PMC 6682551. PMID 31406574.
- Vankadari N (August 2020). “Structure of Furin Protease Binding to SARS-CoV-2 Spike Glycoprotein and Implications for Potential Targets and Virulence”. The Journal of Physical Chemistry Letters. 11 (16): 6655–6663. doi:10.1021/acs.jpclett.0c01698. PMC 7409919. PMID 32787225.
- Coutard B, Valle C, de Lamballerie X, Canard B, Seidah NG, Decroly E (April 2020). “The spike glycoprotein of the new coronavirus 2019-nCoV contains a furin-like cleavage site absent in CoV of the same clade”. Antiviral Research. 176: 104742. doi:10.1016/j.cub.2020.03.022. PMC 7114094. PMID 32057769.
- Zhang T, Wu Q, Zhang Z (April 2020). “Probable Pangolin Origin of SARS-CoV-2 Associated with the COVID-19 Outbreak”. Current Biology. 30 (7): 1346–1351.e2. doi:10.1016/j.cub.2020.03.022. PMC 7156161. PMID 32197085.
- Wu Y, Zhao S (December 2020). “Furin cleavage sites naturally occur in coronaviruses”. Stem Cell Research. 50: 102115. doi:10.1016/j.scr.2020.102115. PMC 7836551. PMID 33340798.
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, et al. (February 2020). “Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding”. Lancet. 395 (10224): 565–574. doi:10.1016/S0140-6736(20)30251-8. PMC 7159086. PMID 32007145.
- O’Keeffe J, Freeman S, Nicol A (21 March 2021). The Basics of SARS-CoV-2 Transmission. Vancouver, BC: National Collaborating Centre for Environmental Health (NCCEH). ISBN 978-1-988234-54-0. Archived from the original on 12 May 2021. Retrieved 12 May 2021.
- Xiao K, Zhai J, Feng Y, Zhou N, Zhang X, Zou JJ, et al. (July 2020). “Isolation of SARS-CoV-2-related coronavirus from Malayan pangolins”. Nature. 583 (7815): 286–289. Bibcode:2020Natur.583..286X. doi:10.1038/s41586-020-2313-x. PMID 32380510. S2CID 218557880.
- Zhao J, Cui W, Tian BP (2020). “The Potential Intermediate Hosts for SARS-CoV-2”. Frontiers in Microbiology. 11: 580137. doi:10.3389/fmicb.2020.580137. PMC 7554366. PMID 33101254.
- “Why it’s so tricky to trace the origin of COVID-19”. Science. National Geographic. 10 September 2021.
- Hu B, Guo H, Zhou P, Shi ZL (March 2021). “Characteristics of SARS-CoV-2 and COVID-19”. Nature Reviews. Microbiology. 19 (3): 141–154. doi:10.1038/s41579-020-00459-7. PMC 7537588. PMID 33024307.
- Giovanetti M, Benedetti F, Campisi G, Ciccozzi A, Fabris S, Ceccarelli G, et al. (January 2021). “Evolution patterns of SARS-CoV-2: Snapshot on its genome variants”. Biochemical and Biophysical Research Communications. 538: 88–91. doi:10.1016/j.bbrc.2020.10.102. PMC 7836704. PMID 33199021. S2CID 226988090.