What is SOS Response or SOS Repair?
- The SOS response, a pivotal cellular mechanism, is activated in response to DNA damage. This intricate system momentarily halts the cell cycle, facilitating DNA repair and mutagenesis. Central to this system is the RecA protein, analogous to Rad51 in eukaryotes.
- When stimulated by single-stranded DNA, RecA plays a crucial role in deactivating the LexA repressor, thereby initiating the SOS response. This repair mechanism, while indispensable, is error-prone and has been linked to DNA alterations across diverse species.
- The foundational understanding of the SOS response can be attributed to the pioneering work of Evelyn Witkin and further elucidated through the collaborative efforts of Witkin and Miroslav Radman, who detailed the bacterial SOS response to UV radiation. Their groundbreaking discovery marked the first elucidation of a coordinated cellular stress response.
- Termed as SOS repair, “bypass” repair, or “emergency” repair, this DNA repair mechanism was introduced to the scientific community in 1975 by Miroslav Radman. Its primary function is to rectify DNA damage resulting from environmental stressors.
- The system is a regulatory nexus, encompassing a myriad of inducer proteins that mend the damaged DNA. A key component of the SOS system is the LexA repressor protein. The RecA protein, omnipresent within the cell, modulates LexA’s activity. Specifically, RecA governs the suppression or expression of the LexA repressor.
- The term “SOS response system” delineates the process wherein an organism triggers the synthesis of the activator protein, RecA. This culminates in the dissociation of the LexA repressor, subsequently activating the SOS inducer proteins.
- Notably, the SOS repair system operates without a DNA template, rendering it susceptible to errors. This article delves into the intricacies of the SOS repair system, elucidating its components, underlying principles, and the mechanisms governing its activation and inactivation.
- In essence, the SOS repair system employs the RecA regulatory protein to inhibit the repressor, subsequently activating the SOS inducer genes to rectify DNA damage. The acronym “SOS” metaphorically signifies a distress call, or “Save Our Soul”. The system remains dormant until the RecA protein is transformed into RecA protease. While it doesn’t offer a comprehensive repair, it bestows the organism with a tolerance to the damage.
- In an undamaged DNA scenario, a bacterial cell has no requirement for the activation of DNA repair genes. This necessitates a regulatory entity to oversee the expression of such genes. LexA fulfills this role, acting as a repressor protein that latches onto specific DNA sites, termed the SOS box, thereby inhibiting the activity of SOS genes.
- However, in the presence of mutated DNA, the deactivation of the LexA repressor becomes imperative to stimulate the expression of SOS genes. Within the SOS system, RecA serves as the catalyst, promoting the degradation of the repressor protein, which in turn facilitates the expression of SOS genes, translating into various DNA repair inducer proteins.
Definition of SOS Response or SOS Repair
The SOS response or SOS repair is a cellular mechanism activated in response to DNA damage, where the cell cycle is temporarily halted to facilitate DNA repair and mutagenesis. Central to this system is the RecA protein, which, when stimulated by single-stranded DNA, deactivates the LexA repressor, initiating the repair process. This system, while essential, is error-prone and can lead to DNA alterations.
Elements of SOS Response or SOS Repair System
The SOS response or SOS repair system is a sophisticated cellular mechanism that orchestrates the repair of DNA damage. This system is composed of a series of elements, each playing a distinct role in the DNA damage response. Here, we delineate the primary components of the SOS system:
- Regulatory Protein (RecA):
- Encoded by: RecA gene
- Function: Acts as the primary regulator of the SOS system. It facilitates the activation of the repressed SOS system by preventing the binding of LexA to the SOS operator.
- Repressor Protein (LexA):
- Encoded by: LexA gene
- Function: Serves as an inhibitor of the SOS system. LexA binds to the operator, leading to the repression or inactivation of the SOS system.
- Inducer Proteins:
- These proteins are encoded by genes associated with the SOS box. Their activation is contingent upon the specific type of DNA damage encountered. Key inducer proteins include:
- uvrA: Repairs short patch nucleotide damage, cross-links, and long patch nucleotide damage.
- uvrD: Addresses cross-links, Me-directed mismatches, double-stranded gaps, and short patch nucleotide damage.
- umuC: Functions to bypass the lesion site in DNA, resulting in mutagenesis.
- umuD: Similar to umuC, it bypasses the lesion site, leading to mutagenesis.
- ruv: Engages in the repair of recombinant DNA damage.
- recN: Repairs recombinant DNA damage and addresses gaps in double-stranded DNA.
- recQ: Specializes in repairing recombinant DNA damage.
- dinA: Bypasses the lesion site in DNA, leading to alterations in the normal DNA sequence.
- sulA: Acts as an inhibitor of cell division, ensuring that damaged DNA is not propagated.
- These proteins are encoded by genes associated with the SOS box. Their activation is contingent upon the specific type of DNA damage encountered. Key inducer proteins include:
In summary, the SOS response system is a coordinated network of proteins, each with specific roles, working in tandem to address DNA damage. This system ensures the integrity of the genetic material, allowing for the proper functioning and survival of the cell.
Functions of SOS Gene Products
The SOS response in bacteria is a coordinated cellular mechanism that is activated in the face of DNA damage. The products of SOS genes play pivotal roles in addressing this damage, ensuring the preservation of genetic integrity.
- Direct DNA Repair Functions:
- Several SOS gene products are directly implicated in DNA repair. Notably:
- Excision Repair Proteins: UvrA and UvrB are involved in the excision repair pathway, which identifies and rectifies damaged DNA segments.
- Daughter-Strand Gap Repair Proteins: RuvA, RuvB, and RecA play crucial roles in repairing gaps in the daughter DNA strand post-replication.
- Several SOS gene products are directly implicated in DNA repair. Notably:
- Indirect DNA Repair Functions:
- SulA Protein: SulA acts as an inhibitor of cell division. By halting cell division while the SOS response is operational, SulA ensures that multiple chromosome copies remain within the same cell. These copies can then serve as templates for the repair of damaged DNA molecules.
- SOS Mutagenesis Proteins: UmuD and UmuC, products of the SOS gene, are central to the SOS mutagenesis process. This mechanism is viewed as a last-resort strategy wherein irreparable DNA damage is converted into a readable sequence, albeit often with errors. Post-synthesis, UmuD undergoes a transformation into its active form, UmuD’. This activation is achieved through a specific proteolytic cleavage, akin to the processes that deactivate LexA and l repressor. However, in this context, the cleavage results in activation. UmuD’ collaborates with UmuC and RecA, hypothesized to restrict the proofreading activity of DNA polymerase III.
- Less Understood Functions:
- Some SOS genes encode functions that remain enigmatic. This ambiguity might arise from the fact that E. coli is frequently studied under laboratory conditions, which might not reflect its natural habitat. In nature, E. coli predominantly resides in environments like the digestive tract or diluted aquatic ecosystems. It is plausible that the strategies enhancing cell survival post-DNA damage in these natural settings differ from those observed under controlled laboratory conditions.
SOS Repair Mechanism
The SOS repair mechanism is an intricate cellular process that the organism employs in response to DNA damage. This mechanism operates in a series of coordinated steps to ensure the integrity of the genetic material:
- Activation of RecA Protein:
- Under conditions of significant DNA damage or stress, the cell activates the RecA protein. This protein patrols the cellular environment, seeking out damaged DNA regions.
- Binding to Single-Stranded DNA:
- RecA specifically associates with single-stranded DNA fragments. Upon binding, it forms a filamentous structure encircling the DNA.
- Interaction with LexA Repressor:
- The nucleoprotein filament, orchestrated by RecA, subsequently interacts with the LexA repressor protein. This interaction transforms RecA into its protease form, termed RecA protease.
- Autocatalytic Proteolysis of LexA:
- The emergence of RecA protease triggers the autocatalytic degradation of the LexA repressor. Consequently, LexA becomes incapable of binding to the SOS operator.
- Activation of Inducer Proteins:
- With LexA inactivated, inducer proteins are activated. These proteins undertake the task of DNA repair. However, it’s noteworthy that this repair process might introduce alterations to the DNA sequence.
- Deactivation of the SOS System:
- Post-repair, RecA’s proteolytic capability diminishes. LexA repressor can then re-associate with the SOS operator, effectively deactivating the SOS system.
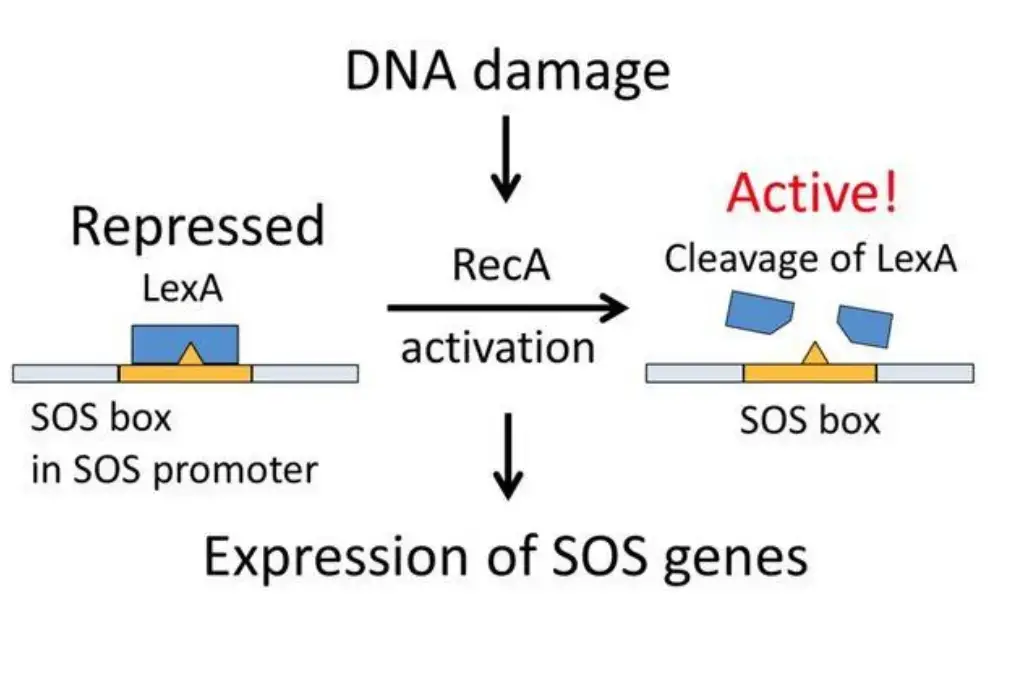
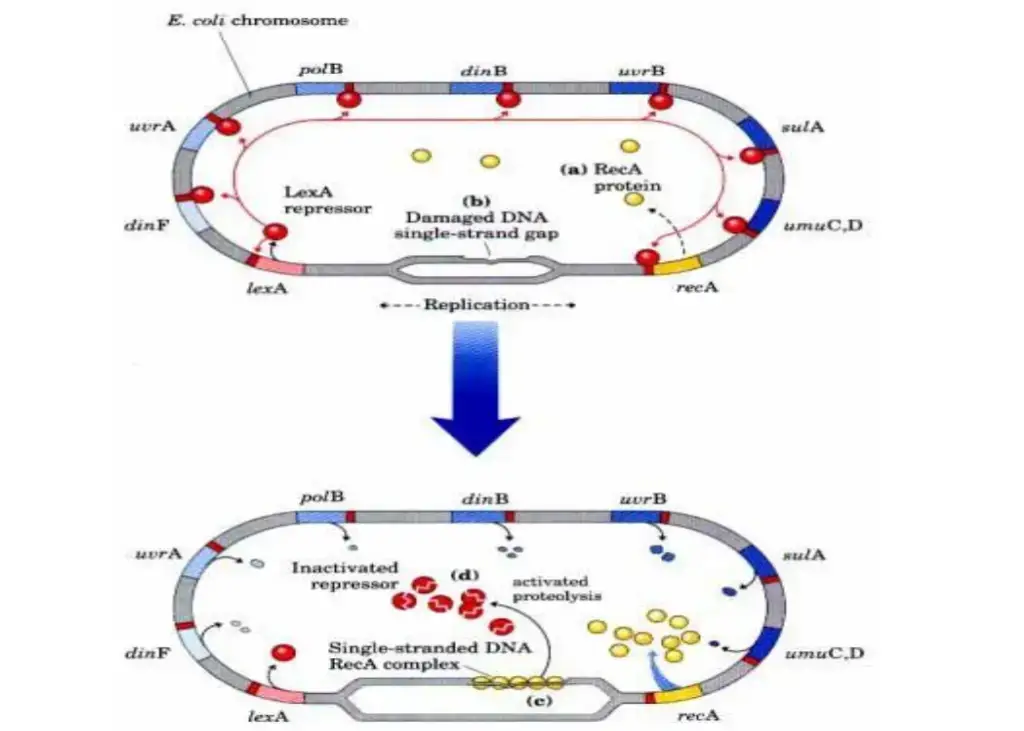
Under typical growth conditions, the SOS genes are under the negative regulation of LexA repressor protein dimers. LexA binds to a specific 20-bp consensus sequence, known as the SOS box, located in the operator region of these genes. The degree of expression of some SOS genes, even in their repressed state, is contingent upon LexA’s affinity for their respective SOS box. DNA damage, particularly the accumulation of single-stranded DNA regions at replication forks, activates the SOS genes. These regions arise when DNA polymerase encounters an impediment. RecA, in an ATP-dependent manner, forms a filament around these regions, becoming activated. This activated RecA facilitates the detachment of LexA repressor from the operator.
As the concentration of LexA diminishes, the repression of SOS genes decreases in accordance with LexA’s affinity for the SOS boxes. This hierarchical activation ensures a sequential activation of repair mechanisms. Genes with weaker SOS boxes are prioritized, initiating nucleotide excision repair (NER) mechanisms. If NER proves insufficient, the concentration of LexA further depletes, leading to the activation of genes with stronger LexA boxes. This includes genes like sulA, which inhibits cell division, and umuD and umuC, which are involved in mutagenic repair. The overall design ensures that the cell can respond to varying degrees of DNA damage, from endogenous levels to more severe damage scenarios.
SOS Response Inactivation and Activation
SOS Inactivation
The SOS system is inherently designed to remain inactive under conditions where the DNA is intact and healthy. This inactivation is primarily orchestrated by the LexA promoter, which synthesizes the LexA repressor protein. This repressor protein binds to a specific consensus sequence, comprising 20 base pairs known as the SOS-box. By doing so, LexA effectively inhibits the SOS system. As a result, the SOS box is occluded by LexA, preventing the activation of SOS genes that are instrumental in repairing damaged DNA.
SOS Activation
Conversely, when DNA integrity is compromised, especially when conventional repair mechanisms are inadequate, the SOS repair system is mobilized. Organisms autonomously activate this system in response to various detrimental factors, including UV-light exposure.
Activation of the SOS system is particularly triggered by extensive DNA damage that results in single-strand breakages at replication forks. Such damage instigates the activation of the RecA regulatory protein, which then associates with the single-stranded DNA, utilizing cellular energy derived from ATP.
The union of RecA protein and single-stranded DNA culminates in the formation of a right-handed nucleoprotein complex, often referred to as the “RecA + ssDNA filament”. This complex subsequently interacts with the LexA repressor, inducing proteolytic cleavage of the LexA dimer. This cleavage event is facilitated by the transformation of the RecA protein into its protease form, effectively neutralizing the inhibitory activity of the LexA protein. With LexA inactivated, genes associated with the SOS box are expressed, giving rise to various inducer proteins tasked with mending the damaged DNA.
It’s noteworthy that the expression of these inducer proteins is not simultaneous but is contingent upon the nature and extent of the DNA damage. The SOS system’s activation and deactivation are, therefore, dynamically regulated, being activated in the presence of the RecA activator protein and deactivated in its absence.
Regulatory Proteins of SOS Response
The SOS response, a cellular mechanism activated in response to DNA damage, is governed by a set of regulatory proteins that play pivotal roles in DNA repair and genetic recombination. Among these, RecA and LexA are the primary regulatory proteins.
1. RecA Protein:
- Function: Beyond its regulatory role, RecA is instrumental in genetic recombination and DNA repair.
- Structure and Activation: Research conducted in vitro suggests that the activated form of RecA manifests as a helical filament. This filament comprises multiple RecA molecules polymerized on single-stranded DNA, in association with ATP or dATP. This structural configuration is indispensable for its roles in recombination and repair.
2. LexA Protein:
- Function: LexA operates as a repressor, akin to the phage l repressor.
- Structure: LexA is characterized by an N-terminal DNA-binding domain and a C-terminal dimerization domain. In its bound state, LexA exists as a dimer, associating with dyad-symmetric DNA sites.
- Proteolytic Cleavage: A unique cleavage site exists between LexA’s two domains. Cleavage at this site decouples the DNA-binding function from the dimerization function, rendering the protein incapable of dimerization and thereby diminishing its DNA-binding efficacy. Notably, this proteolytic cleavage is a self-processing event intrinsic to LexA, with RecA acting as a “co-protease” to stimulate the reaction.
Prophage Induction: In cells harboring a prophage, such as phage l, the CI repressor of the prophage can undergo a cleavage process similar to LexA, driven by active RecA. This process, termed prophage induction, leads to the activation of previously suppressed viral lytic genes, culminating in lytic viral growth and the release of progeny viruses. The CI repressor’s sensitivity to active RecA is believed to be a viral adaptation to cellular signals indicating potential cell demise. The cleavage rate of the l repressor is notably slower than LexA, ensuring that minor DNA damages do not trigger prophage induction. This differential sensitivity between LexA and phage repressors reflects their distinct regulatory environments. While LexA is primed for rapid response to even minor inductions, l repressor is calibrated for a more delayed response, activated only by damage levels threatening cell viability.
Reversibility: A distinguishing feature between the SOS response and prophage induction is their reversibility. The SOS response can be reversed, allowing cells to revert to their normal growth state. In contrast, prophage induction is irreversible due to the intricate regulatory circuitry of the l genetic switch, which suppresses continued CI expression even if cleavage is halted.
In summary, the regulatory proteins of the SOS response, particularly RecA and LexA, play crucial roles in orchestrating the cell’s response to DNA damage, ensuring genetic integrity and cell survival.
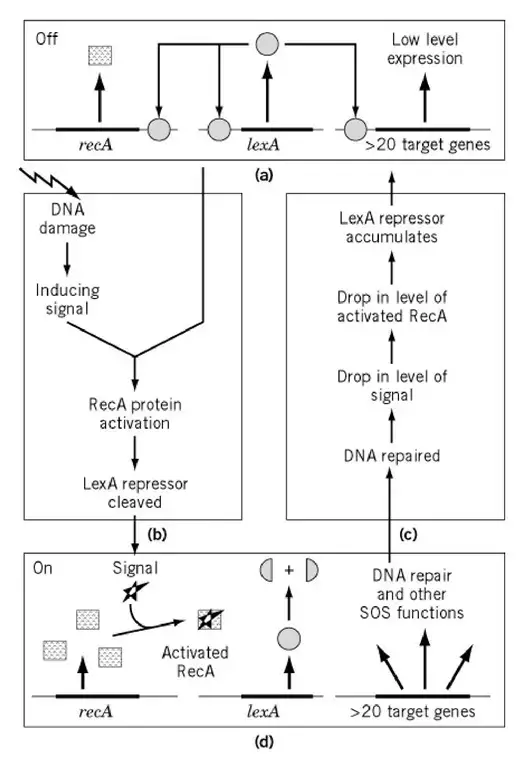
About Above Image: The regulating system SOS. (a) The system’s condition during normal cell proliferation. LexA is an active protein that inhibits the synthesis of RecA (left) and SOS proteins (right). (b). DNA damage induces induction and transition to the induced state. The activated RecA protein causes the LexA repressor protein to self-cleave, rendering it inactive as a repressor. (c). Induced SOS condition In the absence of an active LexA, the recA and SOS genes are highly expressed. Prophage induction occurs if the cell harbours a l prophage and remains in this state for an extended period of time. (c) Transition to the state of normal growth. RecA activation is reversible, hence DNA repair results in deactivation of RecA. The degree of RecA coprotease activity regulates the system’s state and its transitions between two states.
Note: The LexA repressor limits SOS gene expression during normal cell growth. The RecA protein is activated by therapies that induce the SOS response.
| Gene Name | Protien encoded/roel in DNA repair |
| Pol B (din A) | Encoded polymerisation subunit of DNA polymerase II, required for replication restrat in recombinational repair |
| uvrA , uvrB | Encode ABC excinuclease subunit UvrA and UvrB |
| umuC, umuD | Encode DNA polymerase V |
| sulA | Encode protien that iinhibit cell division,possibly to allow time for DNA repair |
| dinB | Encodes DNA polymerase IV |
| uvrD | Encodes DNA helicase II (DNA unwinding protien) |
| recA | Encodes RecA protien rwequired for error-prone repair and recombinational repair |
| ssb | Encodes ssDNA binding protien (SSB) |
| recN | Required for recombinational repair. |
| himA | Encodes subunit of integration host factor,involved in site specific recombination, replication, transposition,regulation of gene expression. |
| dinF | Gene of unknown function |
| dinD | Gene of unknown function |
E. coli SOS System Mechanism
The SOS system in E. coli is a sophisticated cellular response mechanism activated upon DNA damage. This damage can arise from various sources, including cross-linking compounds, UV irradiation, and alkylating chemicals.
- Detection of DNA Damage:
- The RecA protein plays a pivotal role in sensing DNA damage. When DNA is compromised, RecA transforms into its active form, known as LexA protease.
- Activation of RecA:
- Upon detecting broken DNA strands, RecA becomes active and targets its repressor, LexA, for removal. This action ensures that the DNA repair mechanisms are no longer inhibited and can be mobilized to address the damage.
- Autoregulation of the LexA Operon:
- With the elimination of the LexA dimer repressor, the LexA operon gains the capability for autoregulation. This means that the system can modulate its own activity based on the extent of DNA damage and the cellular environment.
- Multifaceted Role of RecA:
- Beyond its role as a LexA protease, RecA is also involved in several specialized DNA processes. These include the annealing of single-stranded DNA and facilitating strand transfer, both of which are crucial for DNA repair and recombination.
- Enhanced DNA Repair Capabilities:
- The activation of the SOS system augments the cell’s DNA repair mechanisms. This encompasses excision repair, post-replication repair, and even enhanced mutagenesis. Additionally, the system can trigger prophage induction, a process where latent viral DNA integrated into the bacterial genome becomes active.
- Cellular Responses:
- Beyond DNA repair, the SOS system can also influence other cellular processes. For instance, it can inhibit cell division, ensuring that damaged DNA is not propagated to daughter cells. Furthermore, the system can modulate cellular respiration, adjusting the cell’s metabolic activity in response to DNA damage.
In essence, the E. coli SOS system is a coordinated and dynamic response to DNA damage, ensuring the preservation of genetic integrity and promoting cell survival in the face of various genotoxic stresses.
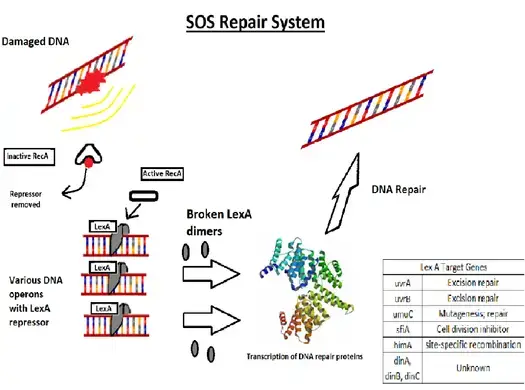
Important Note
The SOS response is a critical cellular mechanism in E. coli and various other bacteria, activated in response to significant DNA damage. This response is orchestrated primarily by two central proteins: LexA and RecA.
- Nature of the SOS Response:
- The SOS response is a global control network activated when the DNA of certain organisms undergoes such extensive damage that conventional repair systems are inadequate to rectify it. Under these circumstances, regular DNA synthesis is halted, necessitating the activation of the SOS response.
- Distribution in the Bacterial Domain:
- While the SOS response is widespread within the Bacteria domain, it is notably absent in certain bacterial phyla, such as the Spirochetes.
- Dependence on RecA and LexA:
- The SOS response, akin to recombination repair, relies heavily on the functions of the RecA and LexA proteins. RecA binds to single- or double-stranded DNA breaks and gaps that arise when DNA synthesis is interrupted. This binding facilitates DNA repair via recombination.
- Activation Mechanism:
- The primary cellular signals prompting the SOS response are segments of single-stranded DNA (ssDNA). These arise from halted replication forks or double-strand breaks and are unwound by DNA helicase. During the initiation phase, RecA protein, in an ATP hydrolysis-driven process, forms RecA–ssDNA filaments. These filaments enhance LexA’s autoprotease activity, leading to LexA dimer cleavage and its subsequent degradation.
- Role of LexA:
- LexA functions as a transcriptional repressor, forming a homodimer that binds to SOS boxes or operator sequences. In E. coli, LexA is known to regulate the transcription of approximately 48 genes, encompassing its own gene and that of RecA. Many genes pivotal for DNA synthesis and repair are negatively regulated by LexA. Its degradation activates genes responsible for excision repair.
- DNA Repair Mechanism:
- MutS plays a role in identifying base mismatches and moves along the DNA. MutL, upon binding to MutS, links it to MutH, necessitating DNA looping for this interaction. MutH discerns the methylated DNA strand, representing the non-mutated parental strand, and facilitates its repair.
In summary, the SOS response is an intricate mechanism ensuring the preservation of bacterial DNA integrity in the face of extensive damage. The coordinated actions of LexA and RecA proteins, along with other associated molecules, ensure the timely repair and maintenance of the bacterial genome.
Importance of SOS Repair
The SOS repair system is a vital cellular response mechanism in bacteria, activated in the face of extensive DNA damage. Its significance can be understood from the following points:
- Preservation of Genetic Integrity:
- The primary objective of the SOS repair system is to maintain the integrity of the bacterial genome. By repairing DNA damage, the system ensures that genetic information is accurately passed on during cell division.
- Response to Various DNA Damages:
- The SOS repair system is versatile and can address a wide range of DNA damages, including those caused by UV radiation, chemical mutagens, and other environmental stressors.
- Global Cellular Response:
- The SOS response is a global cellular mechanism, meaning it coordinates multiple genes and pathways to address DNA damage. This comprehensive response ensures that damage is detected and repaired efficiently.
- Error-Prone Repair:
- While the SOS system is primarily a repair mechanism, it is also error-prone. This means that, in situations where the damage is too severe to be accurately repaired, the system may introduce mutations. While this can lead to errors, it also provides a potential evolutionary advantage by introducing genetic diversity, which might be beneficial under certain environmental conditions.
- Regulation of Cell Division:
- The SOS repair system can temporarily halt cell division, ensuring that damaged DNA is not passed on to daughter cells. This pause allows the cell adequate time to repair the damage before resuming division.
- Survival Mechanism:
- In environments where bacteria face frequent DNA-damaging agents, the SOS repair system enhances bacterial survival. By repairing or bypassing DNA lesions, the system ensures that bacteria can continue to grow and replicate even in adverse conditions.
- Framework for Understanding DNA Repair:
- From a research perspective, the SOS repair system in bacteria like E. coli provides a model to understand DNA damage response and repair mechanisms. Insights gained from studying the SOS system have implications for understanding similar processes in more complex organisms.
In essence, the SOS repair system is crucial for bacterial survival, genome stability, and adaptation to environmental challenges. It represents a sophisticated cellular strategy to counteract the detrimental effects of DNA damage.
Quiz
What is the primary objective of the SOS repair system in bacteria?
a) Protein synthesis
b) RNA transcription
c) Preservation of genetic integrity
d) Energy production
Which protein detects broken DNA and becomes active in the SOS repair system?
a) UmuD
b) LexA
c) SulA
d) RecA
Which protein acts as a repressor in the SOS response system?
a) UvrA
b) LexA
c) RecA
d) UmuC
The SOS repair system is activated in response to which of the following?
a) Protein damage
b) RNA damage
c) DNA damage
d) Cell membrane damage
Which protein in the SOS repair system inhibits cell division?
a) UmuD
b) LexA
c) SulA
d) RecA
The error-prone nature of the SOS repair system can lead to which of the following?
a) Enhanced energy production
b) Introduction of mutations
c) Increased protein synthesis
d) RNA transcription errors
Which protein undergoes a transformation into its active form, UmuD’, in the SOS repair system?
a) UmuC
b) LexA
c) UmuD
d) RecA
Which of the following is NOT a direct function of the SOS repair system?
a) DNA replication
b) Daughter-strand gap repair
c) Excision repair
d) DNA damage detection
In the SOS repair system, the activated form of RecA forms a filament around which type of DNA?
a) Double-stranded DNA
b) Methylated DNA
c) Single-stranded DNA
d) Supercoiled DNA
Which of the following is a direct DNA repair protein in the SOS system?
a) UmuC
b) UvrA
c) SulA
d) LexA
FAQ
What is the SOS repair system?
The SOS repair system is a coordinated cellular response mechanism in bacteria that is activated when there is significant DNA damage.
Why is it called the “SOS” repair system?
The term “SOS” is reminiscent of the international distress signal, indicating an emergency. In this context, it signifies the cell’s emergency response to DNA damage.
Which proteins play a central role in the SOS repair system?
The two principal proteins governing the SOS repair system are LexA and RecA.
How is the SOS repair system activated?
The system is activated in response to extensive DNA damage, such as that caused by UV radiation or chemical mutagens. When DNA damage is detected, the RecA protein becomes active and facilitates the removal of the LexA repressor, leading to the activation of the SOS genes.
Is the SOS repair system error-free?
No, the SOS repair system is error-prone. While it primarily functions to repair DNA, it can sometimes introduce mutations, especially when the damage is too severe to be accurately repaired.
What is the role of the LexA protein in the SOS repair system?
LexA acts as a repressor in the SOS response system. Under normal conditions, it binds to the SOS box, preventing the transcription of SOS genes. When DNA damage is detected, LexA is cleaved and inactivated, allowing the SOS genes to be expressed.
How does the SOS repair system enhance bacterial survival?
The SOS repair system allows bacteria to detect, respond to, and repair DNA damage, ensuring that they can continue to grow and replicate even in adverse conditions.
What happens after the DNA damage is repaired in the SOS response?
Once the DNA damage is repaired, the SOS response is deactivated. The LexA repressor is synthesized again and binds to the SOS box, repressing the SOS genes and returning the cell to its normal state.
Are there any other cellular processes similar to the SOS repair system in other organisms?
While the SOS repair system is primarily observed in bacteria, similar DNA damage response and repair mechanisms exist in eukaryotic cells, though they may involve different proteins and pathways.
Why is the study of the SOS repair system important?
Understanding the SOS repair system provides insights into bacterial DNA repair mechanisms, which can have implications for antibiotic development, understanding bacterial evolution, and studying DNA repair processes in more complex organisms.
References
- Polosina, Y. (2014). DNA Repair☆. Reference Module in Biomedical Sciences. doi:10.1016/b978-0-12-801238-3.02435-1
- Cupples, C. G. (2009). DNA Repair. Encyclopedia of Microbiology, 99–112. doi:10.1016/b978-012373944-5.00070-5
- Maslowska KH, Makiela-Dzbenska K, Fijalkowska IJ. The SOS system: A complex and tightly regulated response to DNA damage. Environ Mol Mutagen. 2019 May;60(4):368-384. doi: 10.1002/em.22267. Epub 2019 Jan 7. PMID: 30447030; PMCID: PMC6590174.
- Whitney, A.K., Weir, T.L. Interaction of caffeine with the SOS response pathway in Escherichia coli . Gut Pathog 7, 21 (2015). https://doi.org/10.1186/s13099-015-0069-x
- Leonard Schärfen, Miloš Tišma, Andreas Hartmann, Michael Schlierf bioRxiv 2020.07.14.201889; doi: https://doi.org/10.1101/2020.07.14.201889
- Harshad GhodkeBishnu P PaudelJacob S LewisSlobodan JergicKamya GopalZachary J RomeroElizabeth A WoodRoger WoodgateMichael M CoxAntoine M van Oijen (2019) Spatial and temporal organization of RecA in the Escherichia coli DNA-damage response eLife 8:e42761.
- Hostetler, Zachary. (2018). A Genetically Encoded Fluorescent Amino Acid Reveals Protein Dynamics Regulating the Bacterial DNA Damage Response.
- Podlesek, Z., & Bertok, D. Ž. (2021). The Escherichia coli SOS Response: Much More than DNA Damage Repair. In (Ed.), Escherichia coli [Working Title]. IntechOpen. https://doi.org/10.5772/intechopen.100353
- https://www.sciencedirect.com/topics/neuroscience/sos-response
- https://biologyreader.com/sos-repair.html
- https://www.brainkart.com/article/The-SOS-Response-in-E–coli_27555/
- http://genesdev.cshlp.org/content/15/4/415/F9.expansion.html
- https://www.cleanpng.com/png-sos-response-dna-repair-repressor-lexa-reca-3322583/
- http://what-when-how.com/molecular-biology/sos-response-molecular-biology/
- https://favpng.com/png_view/sos-response-dna-repair-reca-biology-e-coli-png/Ksssdfzn
- https://blogs.scientificamerican.com/lab-rat/the-sos-response-how-bacteria-deal-with-damaged-dna/
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.