What is Small Interfering RNA (siRNA)?
- Small interfering RNA (siRNA), also known as short interfering RNA or silencing RNA, is a type of double-stranded RNA molecules that function within the RNA interference (RNAi) pathway.
- It inhibits translation, hence inhibiting the expression of certain genes with complementary nucleotide sequences, by degrading mRNA after transcription.
- siRNA is physically and functionally distinct from other types of RNA. Other RNAs are typically single-stranded and composed of a polynucleotide chain. On the opposite side, the siRNA differs.
- The siRNA is a molecule that inhibits gene expression. It is also known as tiny interfering ribonucleic acid or silencing RNA. This implies that it silences genes. The overall process of siRNA-mediated gene silencing is known as RNA interference or siRNA knockdown.
- siRNAs are the tiny double-stranded RNA fragments with a dinucleotide overhang at the 3′ end that degrade mRNA and inhibit protein synthesis.
- Andrew Fire of the Carnegie Institution for Science in Washington, D.C., and Craig Mello of the University of Massachusetts in Worcester discovered the RNA interference (RNAi) process in 1998 while studying gene expression in Caenorhabditis worms. In 2006, they were awarded the Nobel Prize for their work with RNAi.
- In 1999, Science reported the discovery of siRNAs and their function in post-transcriptional gene silencing (PTGS) in plants by the group of David Baulcombe at the Sainsbury Laboratory in Norwich, England.
Properties of siRNA
- siRNA is a non-coding double-stranded RNA molecule.
- It is also known as short interfering RNA and silencing RNA.
- Comparable to microRNA (miRNA), its structure is brief and well-defined, typically between 20 and 24 base pairs.
- Their 3′ and 5′ ends are hydroxylated and phosphorylated, respectively.
- The Dicer enzyme is responsible for catalysing the synthesis of siRNA.
- It is utilised to control gene expression by transcriptional or translational repression, making it a potent tool in drug targeting and therapies development.
- A synthetic siRNA with a complementary sequence could theoretically silence any gene. This makes them an essential tool for medication targeting and gene function validation.
- Numerous significant studies have been conducted on the application of siRNA to various fields of medical study.
Structure of siRNA
Naturally occurring siRNAs have a well-defined structure consisting of phosphorylated 5′ ends, hydroxylated 3′ ends, and two overhanging nucleotides.
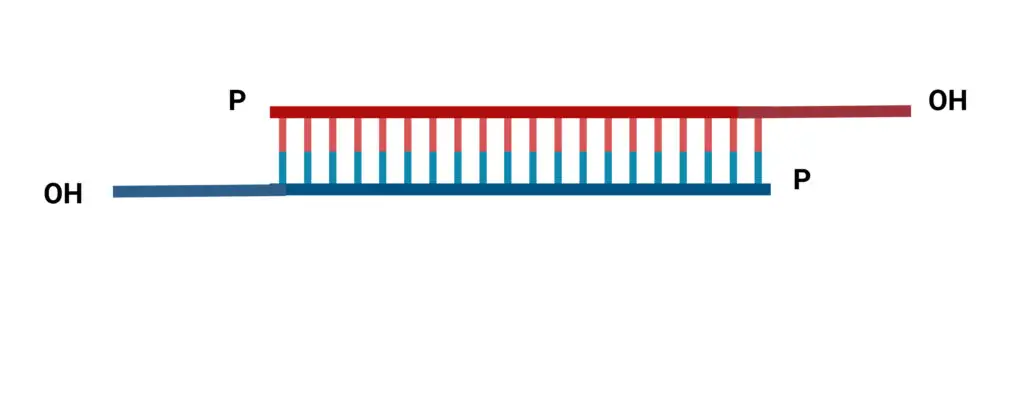
- The siRNA is double-stranded, 20 to 25 nucleotides in length, and double-stranded. The origin of siRNA is exogenous, and it inhibits protein translation.
- In addition to these characteristics, one of the distinguishing features of siRNA is the presence of the 3′ OH dinucleotide overhang.
- It is a dsRNA that is shorter in length and has an overhang at one end.
- One strand of the double-strand is known as the guided strand and the other as the passenger strand. It is also known as a sense strand and an antisense strand.
- David Baulcombe and colleagues described the involvement of siRNA in post-transcriptional modification in 1999.
- It is made up of Adenine, Uracil, Cytosine, and Guanine at the molecular level.
- The phosphodiester bond connects two neighbouring nucleotides, while hydrogen bonds connect two nucleotides from separate strands.
- The 3′ ends of both dinucleotides lack hydrogen bonds.
- From long dsRNAs and tiny hairpin RNAs, the Dicer enzyme catalyses the creation of siRNAs.
- siRNAs can be injected into cells through transfection as well. In the post-genomic age, siRNAs are an essential tool for confirming gene activity and therapeutic targeting, as any gene in principle can be silenced by a synthetic siRNA with a complementary sequence.
Mechanism of siRNA action
The following describes the process by which natural siRNA induces gene silencing via suppression of translation.
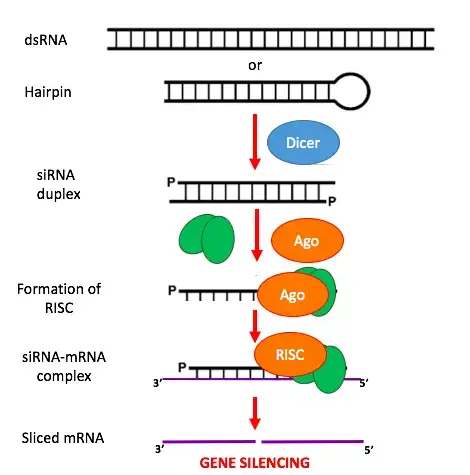
- Dicer is an endoribonuclease that cleaves long dsRNA, which may originate from hairpin, complementary RNAs, or RNA-dependent RNA polymerases. This allows the molecules to form the RNA-Induced Silencing Complex (RISC).
- Once siRNA reaches the cell, it fuses with other proteins to form the RISC.
- Once siRNA is incorporated into the RISC complex, it is unravelled to generate single-stranded siRNA.
- The RISC-complex retains the strand that is thermodynamically less stable due to the base pairing at its 5′ end.
- The RISC complex’s single-stranded siRNA may now scan for and locate a complementary mRNA.
- Once the siRNA (a component of the RISC complex) attaches to its target mRNA, it triggers mRNA cleavage.
- The mRNA is now cut and the cell recognises it as aberrant. This results in the breakdown of the mRNA, preventing its translation into amino acids and then proteins. Consequently silencing the gene that codes for this mRNA.
siRNA is also similar to miRNA; however, miRNAs are derived from shorter stemloop RNA products, typically silence genes by repressing translation, and have a broader specificity of action, whereas siRNAs typically function by cleaving the messenger RNA (mRNA) prior to translation, and have 100% complementarity, thus very tight target specificity.
Intracellular delivery of siRNA
Intracellular delivery of siRNA continues to be a difficulty. There are three primary delivery methods for siRNA that vary in their efficacy and toxicity.
Transfection
- In this technique, siRNA against the target gene must first be designed.
- Once the siRNA has been designed to target a specific gene, it must be effectively delivered using a transfection protocol.
- Typically, cationic liposomes, polymer nanoparticles, and lipid conjugation are utilised for delivery.
- This method is advantageous because it can deliver siRNA to the majority of cell types, is highly effective and reproducible, and is commercially available.
- Lipofectamine and Neon Transfection are the most commonly used commercial transfection agents for siRNA.
- However, it is not compatible with all cell types and is inefficient in vivo.
Electroporation
- Electrical pulses are also used to deliver siRNA into cells intracellularly.
- Phospholipids compose the cell membrane, making it susceptible to an electric field.
- When rapid but powerful electrical pulses are initiated, lipid molecules undergo phase transitions due to heating and reorient themselves.
- This causes the formation of hydrophilic pores and localised perturbations in the lipid bilayer cell membrane, resulting in a temporary loss of semipermeability.
- This permits the escape of numerous intracellular constituents, such as ions and metabolites, and the simultaneous uptake of drugs, molecular probes, and nucleic acids.
- Electroporation is advantageous for difficult-to-transfect cells, but cell death is more likely with this technique.
- This method was used to deliver VEGF-targeting siRNA into xenografted tumours in nude mice, resulting in a significant decrease in tumour growth.
Viral-mediated delivery
- The gene-silencing effects of transfected siRNA are typically transient, but this difficulty can be circumvented by employing an RNAi strategy.
- This siRNA can be delivered from DNA templates via multiple recombinant viral vectors based on retrovirus, adeno-associated virus, adenovirus, and lentivirus.
- This virus is the most effective at delivering siRNA to target cells because it can transduce nondividing cells and directly target the nucleus.
- These specific viral vectors have been synthesised to effectively facilitate the transfection of siRNA into cells that is not viable on its own.
- In some instances, synthetic viral vectors can integrate siRNA into the cell genome, allowing for stable siRNA expression and long-term gene silencing.
- This method is advantageous because it is in vivo and effective for transfecting cells that are difficult to transfect.
- However, it can induce antiviral responses in certain cell types, resulting in mutagenic and immunogenic effects.
- This technique may be used to silence genes in the central nervous system to treat Huntington’s disease.
Applications of siRNA
- This system is present in almost all eukaryotes and functions to combat viral infections. Today, scientists utilise this understanding for therapeutic gene silencing and gene expression regulation.
- Currently, scientists are manufacturing siRNA molecules that are unique to the mRNA of a gene they desire to inhibit.
- The siRNA can be injected into the cell utilising viral vector-based or nonviral vector-based artificial techniques of gene transfer.
- This technique eliminates the target messenger RNA and controls protein production.
- Researchers are currently attempting to use siRNA-mediated gene silencing to inhibit cancer-causing genes.
- In the gene knockout and gene knockdown techniques, siRNA-mediated gene expression suppression is utilised.
- It is also utilised for target validation, meaning that the investigated target gene can be validated.
- It is also utilised in the investigation and identification of pathways such as cytokinesis, insulin signalling, and cell defence mechanism, among others.
- In addition, it can be used for gene redundancy and gene functional research.
- Brain medication delivery utilises carbon-based and non-carbon-based nanoparticle-mediated siRNA treatment.
Challenges
However, there are obstacles associated with siRNA use. Occasionally, cleavage fails due to mismatches between the siRNA and regions of the target mRNA close to the cleavage site. There are more nonspecific effects of siRNA.
Intersections between RNAi and other pathways result in the occasional activation of these nonspecific effects. Problems include:
- Mammalian cells mount an immunological response after mistaking double-stranded RNA such as siRNA for viral byproducts.
- The modification of the thermodynamic characteristics of siRNA, resulting in a loss of single nucleotide specificity.
- Unintended misdirection. This happens as a result of the unintentional downregulation of genes with incomplete complementarity. This results in concerns with data interpretation and possibly toxicity.
- Too many siRNAs are injected, activating the innate immunological responses of the host. There is evidence that this is caused by the activation of PKR, a dsRNA sensor, as well as other responses.
In recent years, methods to alleviate these negative effects have been discovered. For instance, off-targeting can be mitigated in part by creating appropriate control trials and algorithms that generate off-target-free siRNAs. Genome-wide expression analysis, such as microarray technologies, can be utilised to further enhance algorithms.
In addition, intracellular transport of siRNA continues to provide difficulties. Transfection, electroporation, and viral-mediated delivery are prevalent delivery techniques. Although it is not suitable with all cell types and has limited in vivo efficacy, transfection is the most extensively utilised of these techniques. Ongoing study is being conducted to see how these delivery techniques might be improved.
Despite the fact that intracellular delivery presents obstacles and that siRNA exhibits poor stability and pharmacokinetics, there is a growing interest in the therapeutic applications of the technology. The pharmaceutical industry has made substantial investments in the study and development of siRNA therapeutics for a wide range of medical ailments and disorders.
Due to their modest size of approximately 20 base pairs, siRNAs can pass through parts of the body where larger genetic therapies cannot. This includes the blood-brain barrier, which has historically provided substantial drug delivery issues.
References
- Kamaruzman, Nur & Abd Aziz, Noraini & Poh, Chit & Chowdhury, Ezharul. (2019). Oncogenic Signaling in Tumorigenesis and Applications of siRNA Nanotherapeutics in Breast Cancer. Cancers. 11. 632. 10.3390/cancers11050632.
- Kumar, M., DeVaux, R. S., & Herschkowitz, J. I. (2016). Molecular and Cellular Changes in Breast Cancer and New Roles of lncRNAs in Breast Cancer Initiation and Progression. Molecular and Cellular Changes in the Cancer Cell, 563–586. doi:10.1016/bs.pmbts.2016.09.011
- Dana H, Chalbatani GM, Mahmoodzadeh H, Karimloo R, Rezaiean O, Moradzadeh A, Mehmandoost N, Moazzen F, Mazraeh A, Marmari V, Ebrahimi M, Rashno MM, Abadi SJ, Gharagouzlo E. Molecular Mechanisms and Biological Functions of siRNA. Int J Biomed Sci. 2017 Jun;13(2):48-57. PMID: 28824341; PMCID: PMC5542916.
- Acc. Chem. Res. 2012, 45, 7, 1014–1025 Publication Date:March 13, 2012 https://doi.org/10.1021/ar2002254
- Tuschl, T. (2001). RNA Interference and Small Interfering RNAs. ChemBioChem, 2(4), 239–245. doi:10.1002/1439-7633(20010401)2:4<239::aid-cbic239>3.0.co;2-r
- https://www.astrazeneca.com/r-d/next-generation-therapeutics/small-interfering-rna.html#!
- https://www.news-medical.net/life-sciences/What-is-SiRNA.aspx
- https://mcmanuslab.ucsf.edu/node/268
- https://www.dovepress.com/small-interfering-rnas-sirnas-in-cancer-therapy-a-nano-based-approach-peer-reviewed-fulltext-article-IJN
- https://en.wikipedia.org/wiki/Small_interfering_RNA
- https://www.creative-biolabs.com/gene-therapy/category-small-interfering-rna-sirna-362.htm
- https://geneticeducation.co.in/sirna-small-interfering-rna-structure-and-function/