What is RNA splicing?
In order for mRNA to be translated into a protein, RNA splicing removes the intervening, non-coding sections of genes (introns) from pre-mRNA and links the protein-coding sequences (exons) together.
- RNA splicing is the procedure by which newly created pre-mRNA, also known as hnRNA (heterogeneous nuclear RNA), is processed and mature mRNA is produced.
- hnRNA is processed and transformed to mRNA in the nucleus, which is then transported to the cytoplasm for translation or protein production. It is an alteration after transcription.
- Two scientists, Philip Allen Sharp and Richard J. Roberts, were awarded the 1993 Nobel Prize in Physiology or Medicine for their discovery of RNA splicing.
- In prokaryotes such as bacteria, newly transcribed RNA is immediately available for translation, and both processes can occur simultaneously in the mRNA.
- The majority of eukaryotic genes are transcribed as pre-mRNA and require processing prior to protein synthesis.
- In the process of RNA splicing, the non-coding intervening portions known as ‘introns’ are eliminated and the coding regions known as ‘exons’ are connected.
- RNA splicing is catalysed by spliceosome. Ribozymes (RNA enzymes) facilitate their own splicing.
- Moreover, 5′ capping with the modified Guanine nucleotide and tailing with Poly-A (Adenylate) residues at the 3′ end are performed to protect the coding regions and provide stability to the mature mRNA.
Facts of RNA splicing
- The definition of’splice’ is to join.
- Introns are eliminated and joined to exons.
- Typically found in eukaryotes.
- Exon and Intron both exist in a distinct manner.
- Every molecule begins and concludes with an exon.
- Splicing is the process of removing the introns from the pre-MRNA and linking the exons together.
- It occurs in the nucleus prior to the export of mature MRNA into the cytoplasm.
- In the majority of genes, protein-coding information is interrupted by noncoding sections known as “introns.” The coding sequences are referred to as “exons.”
- Introns: non-coding sequences.
- Exons: coding sequences.
What is Intron?
- Introns are non-protein-coding nucleotide regions in DNA and RNA that are deleted during the precursor messenger RNA (pre-mRNA) stage of mRNA maturation through RNA splicing.
- Introns can range in size from tens to thousands of base pairs and can be found in a variety of genes that produce RNA in the majority of living species, including viruses.
- Introns are noncoding DNA sequences contained within a gene that are deleted during the development of the RNA transcript by the process of RNA splicing.
- The term intron is derived from intragenic region and intracistron, which refer to a stretch of DNA situated between two exons of a gene.
- Both the DNA sequence within a gene and the matching sequence in the unprocessed RNA transcript are referred to as introns.
- RNA splicing removes introns as part of the RNA processing mechanism, either quickly after or simultaneously with transcription.
- Introns are present in the genes of the vast majority of organisms and viruses.
- They can be found in a variety of genes, including as those that produce proteins, ribosomal RNA (rRNA), and transfer RNA (tRNA).
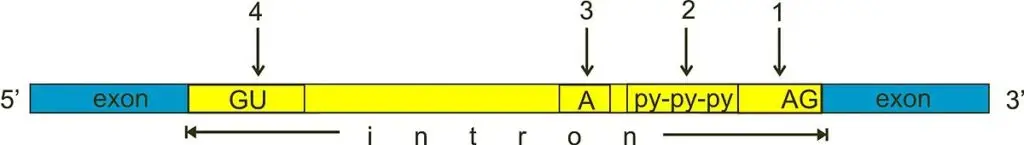
- For splicing to occur within introns, a donor site (5′ end of the intron), a branch site (near the 3′ end of the intron), and an acceptor site are necessary.
- At the 5′ end of the intron, the splice donor site has a nearly constant GU sequence inside a broader, less conserved region.
- The splice acceptor site at the 3′ end of the intron concludes the intron with a sequence of AG that is nearly constant.
- Upstream (5′-ward) from the AG there is a region high in pyrimidines (C and U), or polypyrimidine tract.
- The branchpoint further upstream from the polypyrimidine tract contains an adenine nucleotide important in lariat formation.
- The consensus sequence for an intron is G-G-[cut] (in IUPAC nucleic acid notation).
- -G-U-R-A-G-U (donor site) … intron sequence … Y-U-R-A-C… Y-rich-N-C-A-G-[cut] (branch sequence 20-50 nucleotides upstream of acceptor site). -G (acceptor site).
- The exact sequence of intronic splicing elements and the number of nucleotides between the branchpoint and the closest 3′ acceptor site affect splice site selection, it is noted.
- In addition, point mutations in the underlying DNA or transcriptional mistakes can trigger a cryptic splice site in a normally unspliced region of the transcript. This produces mature messenger RNA without a portion of one exon.
- Thus, a point mutation, which would ordinarily influence only a single amino acid, can show in the final protein as a deletion or truncation.
What is Exon?
- American biochemist Walter Gilbert coined the term exon in 1978: “The notion of the cistron… must be replaced by that of a transcription unit containing regions that will be lost from the mature messenger – which I suggest we call introns (for intragenic regions) – alternating with regions that will be expressed – exons.”
- Exons are DNA sequences that code for proteins and contain the codons or genetic information necessary for protein production.
- In addition to protein-coding sequences, exons often comprise the 5′- and 3′ untranslated sections of messenger RNA, which contain start and stop codons.
- Exons are present in all creatures, including vertebrates with jaws, yeasts, bacteria, and even viruses.
- Exons are critical for protein production because they contain areas containing codons that code for different proteins.
- An exon is any portion of a gene that will be included in the mature RNA generated by that gene once the introns have been eliminated by RNA splicing.
- Exon refers to both the DNA sequence within a gene as well as the associated RNA transcript sequence.
- Introns are deleted and exons are covalently linked during RNA splicing as part of the process of producing mature RNA. In the same way that the complete set of genes for a species comprises the genome, the complete set of exons comprises the exome.
- Although unicellular eukaryotes such as yeast have no or extremely few introns, metazoan and particularly vertebrate genomes contain a substantial amount of noncoding DNA. For example, in the human genome there are only 1 75% of the genome consists of intergenic DNA, while just 1% of the genome is comprised of exons.
- This can provide a practical advantage in omics-assisted health care (such as precision medicine) because commercial whole exome sequencing is a smaller and less costly issue than commercial whole genome sequencing.
- The significant difference in genome size and C-value among life forms has provided an intriguing problem known as the C-value mystery.
- In 2002, eukaryotic genes in GenBank contained, on average, 5.48 exons per protein-coding gene. The typical exon coded for 30-36 amino acids.
- While the longest exon in the human genome is 11555 base pairs in length, other exons have been discovered to be only 2 base pairs in length.
- A single-nucleotide exon has been identified in the genome of Arabidopsis. Similar to protein-coding mRNA, the majority of noncoding RNA likewise has several exons in humans.
- The exons of protein-coding genes contain the protein-coding sequence as well as the 5′- and 3′-untranslated regions (UTR).
- Some genes contain exons including only portions of 5′-UTR or (less frequently) 3′-UTR, i.e. the UTRs may contain introns. Typically, the first exon has both the 5′-UTR and the initial portion of the coding sequence.
- Exons and introns are present in some non-coding RNA transcripts.
- Alternative splicing can remove distinct introns from the pre-mRNA, allowing mature mRNAs coming from the same gene to include different exons.
- Exonization is the emergence of a new exon as a result of intron mutations.
What is Spliceosome?
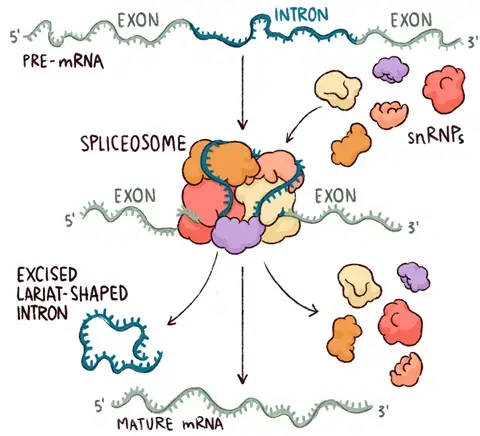
- The spliceosome is a vast complex or enormous molecular mechanism that performs RNA splicing.
- It is a huge and intricate molecular machine that removes introns from the pre-Mrna molecule.
- The interaction of RNAs and these proteins produces an RNA-protein complex known as small nuclear ribonucleoproteins (snRNPs).
- These are predominantly limited to the nucleus, where they continue to be associated with immature pre-RNA transcripts.
- The spliceosome acts as an editor that removes unneeded and erroneous material (introns) to produce a viable final cut.
- All spliceosomes participate in the elimination of introns and the ligation of remaining exons.
- Tiny nuclear ribonucleoproteins are complexes of small nuclear RNA and proteins (snRNP, pronounced “snrnps”). There are five U1, U2, U4, U5, and U6 snrnps.
Types of Spliceosome
There are two types of spliceosomes:
- Major Spliceosomes – They are responsible for 99.5% of intron elimination. They are composed of the snRNPs U1, U2, U4, U5, and U6.
- Minor Spliceosomes – They are responsible for the elimination of 0.5% of introns. They are composed of the snRNPs U11, U12, U4atac, U5, and U6atac.
RNA Splicing Process/ Mechanism
RNA splicing is an important step in gene expression since it is responsible for eliminating non-coding portions known as introns from the parent RNA transcript and putting together coding areas known as exons to form a mature mRNA molecule. Let’s take a step-by-step look at the RNA splicing process:
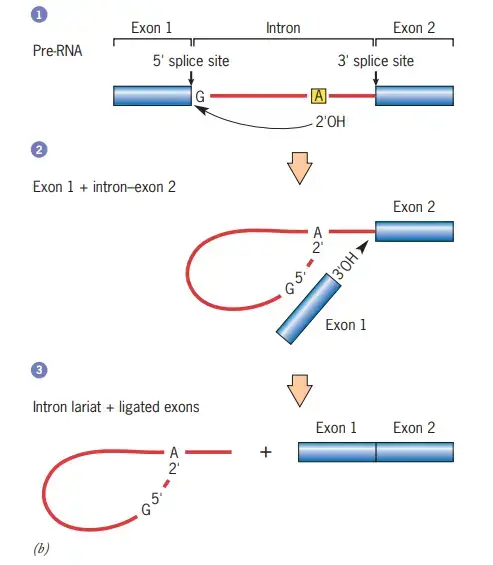
- Binding of Spliceosomes: The process begins with ribonucleoproteins or spliceosomes binding to the introns at the splice site. Spliceosomes are huge molecular complexes that contain RNA as well as proteins.
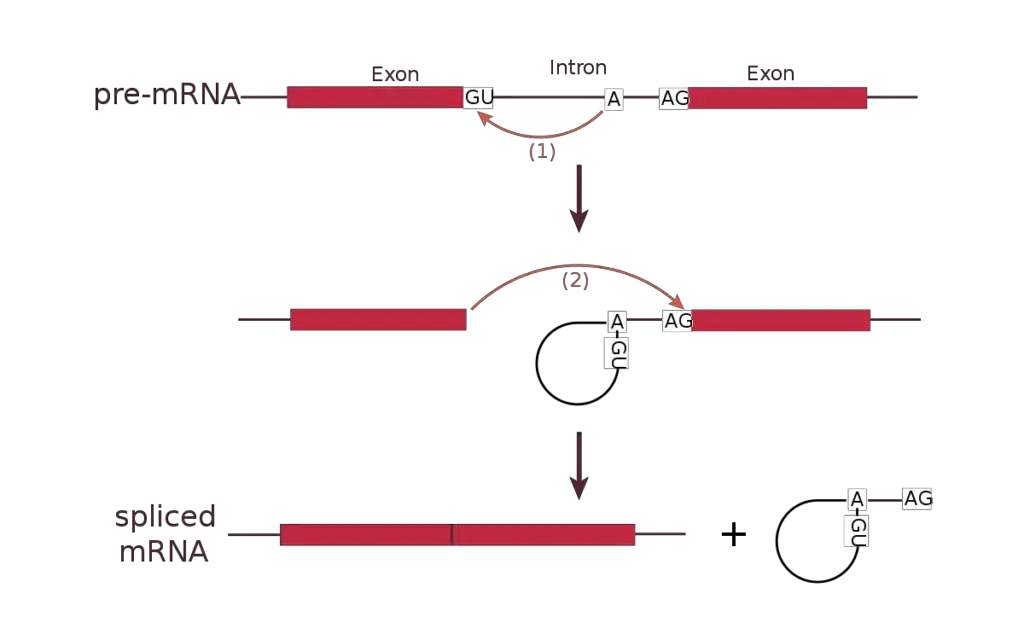
- Transesterification Reaction: Spliceosome binding causes a biological reaction known as transesterification. A certain nucleotide on the intron, determined during spliceosome construction, has a 3’OH group that acts as a nucleophile during this process.
- Nucleophilic Attack at the 5′ Splice Site: The intron nucleotide’s 3’OH group launches a nucleophilic attack on the intron’s first nucleotide at the 5′ splice site. This folds the 5′ and 3′ ends, forming a loop shape. Furthermore, the neighbouring exons are brought closer together.
- Detachment of the Looped Intron: Spliceosomes aid in the separation of the looped intron from the RNA sequence. The intron is removed from the primary transcript while the exons remain intact.
- Ligation of Adjacent Exon Segments: After the intron is removed, a second transesterification reaction ensues. At the 3′ splice site, the 3’OH group of the liberated 5′ exon initiates an electrophilic attack on the nucleotide just downstream of the intron.
- Binding and Intron Segment elimination: The electrophilic assault causes the two neighboring exon segments to bind, resulting in the elimination of the intron segment. This is referred to as ligation.
- Intron Post-Splicing Functions: Introns were previously thought to be “junk” units, but new research has demonstrated their participation in a variety of protein-related functions even after they have been removed during splicing. This demonstrates the importance of introns beyond their involvement in RNA splicing.
- Ribozymes: In addition to spliceosomes, ribozymes, a group of protein/enzymes, play a function in controlling and regulating the splicing process. Ribozymes are RNA molecules that have catalytic activity and can perform particular chemical reactions.
The RNA splicing process, which includes spliceosome binding, transesterification processes, exon ligation, and ribozyme participation, guarantees that introns are removed and exons are properly joined, resulting in mature mRNA molecules for protein synthesis.
Splicing Types
1. Group I and Group II introns Splicing – self-splicing
- Some nuclear, mitochondrial, and chloroplast genes that code for rRNAs, mRNAs, and tRNAs have Group I introns.
- In fungus, algae, and plants, Group II introns are typically present in the main transcripts of mitochondrial or chloroplast mRNAs.
- In bacterium, group I and group II introns are also among the few examples of introns.
- For splicing, neither class requires a high energy cofactor (such as ATP).
- The splicing methods of both groups require two transesterification reaction stages in which a ribose 2′- or 5′-hydroxyl group makes a nucleophilic assault on a phosphorus and a new phosphodiester bond is generated at the expense of the old, thereby maintaining the energy balance.
- These processes resemble the DNA cleavage and reassembly reactions facilitated by topoisomerases and site-specific recombinase.
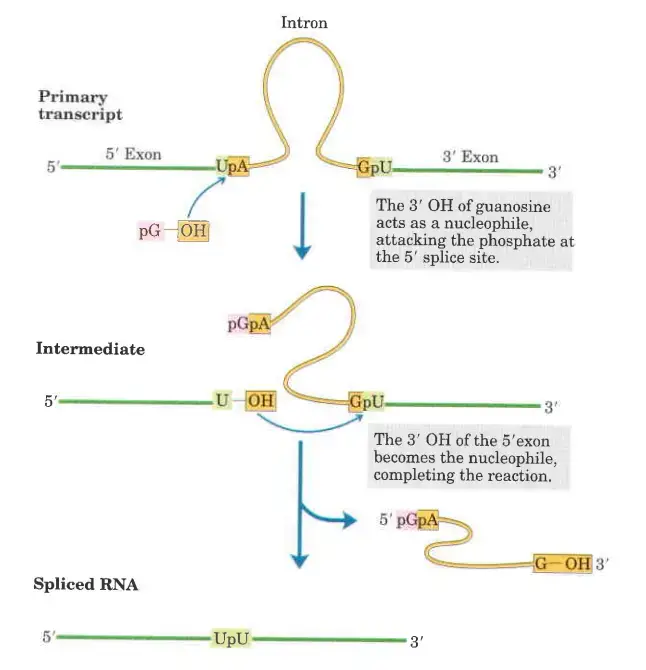
- In the first step of the splicing pathway, the 3′-hydroxyl group of guanosine is utilised as a nucleophile.
- The 3′-hydroxyl group of guanosine forms a standard 3′,5′-phosphodiester connection with the 5′ terminus of the intron.
- The 3′, hydroxyl of the exon that is displaced in this step works as a nucleophile in a comparable reaction that occurs at the 3′ end of the intron. Excision of the intron and ligation of the exons are the end result.
- Introns of group II follow a similar reaction pattern, with the exception of the nucleophile in the initial step, which is the 2′-hydroxyl group of an A residue within the intron. As an intermediary construction, a branched lariat is produced.
2. Alternative Splicing
- Alternative splicing, also known as alternative RNA splicing or differential splicing, is an alternative splicing process that permits a single gene to encode for several proteins.
- Certain exons of a gene may be included or excluded from the final, processed messenger RNA (mRNA) produced from that gene during this procedure. This indicates that the exons are connected in different combinations, resulting in different mRNA strands (alternatives).
- Therefore, proteins translated from alternatively spliced mRNAs will have different amino acid sequences and, frequently, different biological activities.
- Alternative splicing happens following the formation of primary mRNA from DNA. This is known as transcription, as the languages of RNA and DNA are almost identical.
- Both depend on four nucleotide bases. When a ribosome reads this language, it translates it into the language of proteins, which is composed of approximately 21 amino acids.
- Before a primary mRNA can be translated into a protein, it must therefore undergo modification and editing. During normal splicing, a protein and RNA complex known as the spliceosome binds to the primary mRNA.
- The primary mRNA is composed of distinct sections known as introns and exons. These sections are combined, and the introns must be eliminated in order to produce a functional protein.
- The spliceosome is equipped specifically to eliminate introns. Four distinct subunits known as small nuclear ribonucleoproteins (snRNP or “snurp”) compose spliceosomes. Each “snurp” contains two tiny nuclear RNA molecules (snRNAs).
- These RNA strands include sequences of nucleotides that match and bind to certain sites in the exons. The protein part of the spliceosome then functions as an enzyme, eliminating introns and joining exons. This spliced mRNA is now prepared for protein translation.
- Nonetheless, alternative splicing is also possible. While the mechanism as a whole is not well understood, it is known that certain chemical substances can trigger the spliceosome to function in various ways.
- A signal may be used to omit a single exon or many exons from the final mRNA. Other signals and pathways can induce the spliceosome to leave introns undamaged or to bypass substantial portions of the protein.
- Proteins serve a variety of functions in our bodies, and the same DNA blueprint can frequently be used to produce multiple proteins. Below is a chart illustrating the various ways in which a spliceosome might alternatively splice a primary RNA.
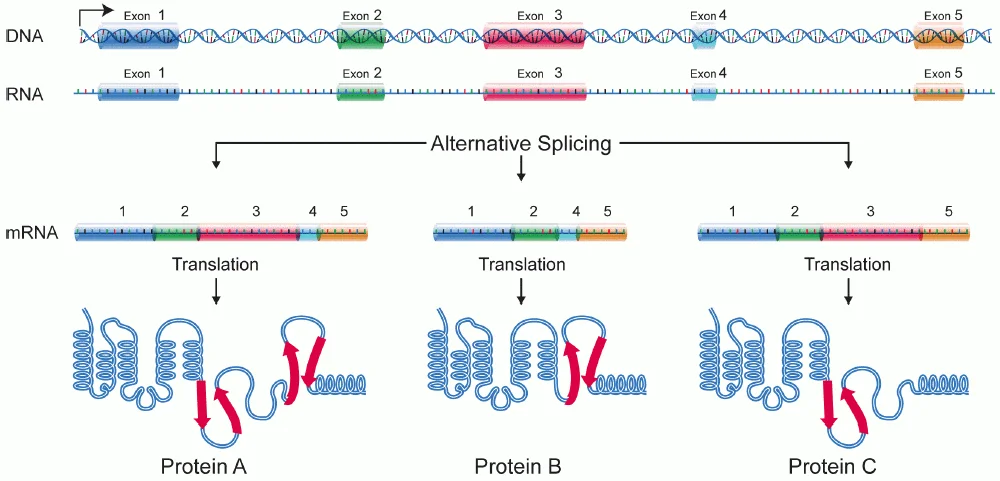
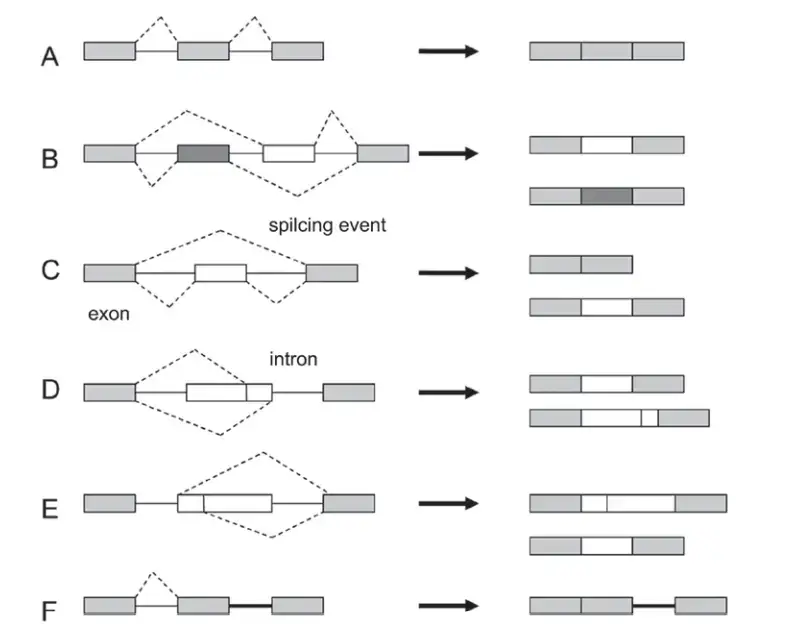
Types of alternative splicing
Five main types of alternative splicing events are depicted.
- Constitutive splicing;
- mutually exclusive exons;
- cassette alternative exon;
- alternative 3′ splice site;
- alternative 5′ splice site; and
- intron retention
Trans Splicing
- Trans splicing is an additional type of alternative splicing in which exons from two distinct genes are assembled by a spliceosome.
- This genetic mechanism has only been observed in a few of unicellular creatures, but it may help explain their genetic variety in the absence of sexual reproduction.
- While sexually reproducing creatures must mate to develop new types, these organisms can do so considerably more quickly.
- This method of gene splicing can quickly produce totally new functionalities in these species, which may be advantageous.
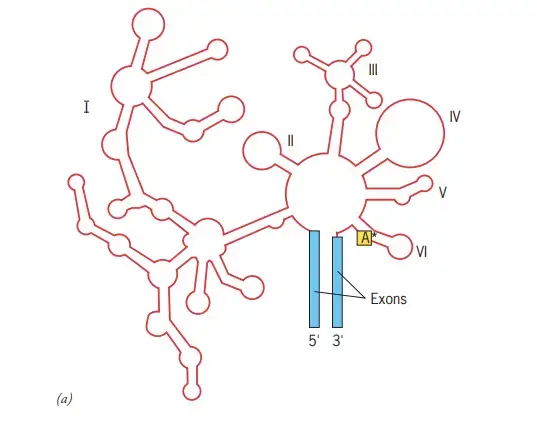
Describe The structure and self-splicing pathway of group II introns in Detail?
- Group II intron structure in two dimensions (shown in red).
- The intron is composed of six distinct domains that radiate from a core structure.
- The asterisk represents the adenosine nucleotide that protrudes from domain VI and creates the lariat structure described in the text.
- The proximity of the two intron–exon borders indicates that the two ends of the intron approach one another.
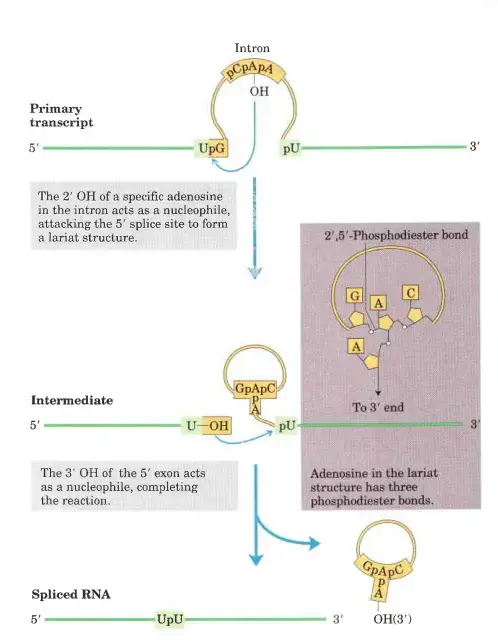
Steps in the self-splicing of group II introns
- In step 1, the 2 OH of an adenosine within the intron (asterisk in domain VI of component a) performs a nucleophilic attack on the 5 splice site, thereby cleaving the RNA and establishing an unusual 2–5 phosphodiester bond with the first nucleotide of the intron.
- This structure with branches is known as a lariat.
- In step 2, the relocated exon’s free 3 OH assaults the 3 splice site, cleaving the RNA at the other end of the intron.
- This reaction results in the release of the intron as a free lariat and the ligation of the 3 and 5 ends of the two flanking exons (step 3).
- The splicing of introns from pre-mRNAs follows a similar mechanism, but unlike self-splicing, these stages require the assistance of a number of extra components.
Schematic model of the assembly of the splicing machinery and some of the steps that occur during premRNA splicing
- In Step 1, the pre-mRNA segment to be spliced is identified.
- At the 5 splice site of the intron, the first splicing component, U1 snRNP, is joined in step 2.
- U1 snRNA is complementary to the 5 splice site of the pre-mRNA, and there is evidence that U1 snRNP initially binds to the 5 side of the intron by forming particular base pairs between the splice site and U1 snRNA (see inset A).
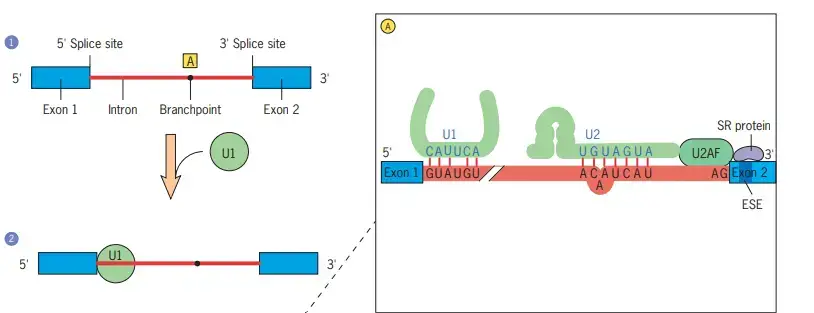
- The U2 snRNP binds to the pre-mRNA (shown in inset A) in a manner that causes a specific adenosine residue (dot) to protrude from the surrounding helix (step 3).
- This location will eventually become the lariat’s branching point. The protein U2AF, which binds to the polypyrimidine tract near the 3 splice site, is believed to recruit U2.
- Additionally, U2AF interacts with SR proteins that bind exonic splicing enhancers (ESEs). These interactions are crucial for detecting intron/exon boundaries.
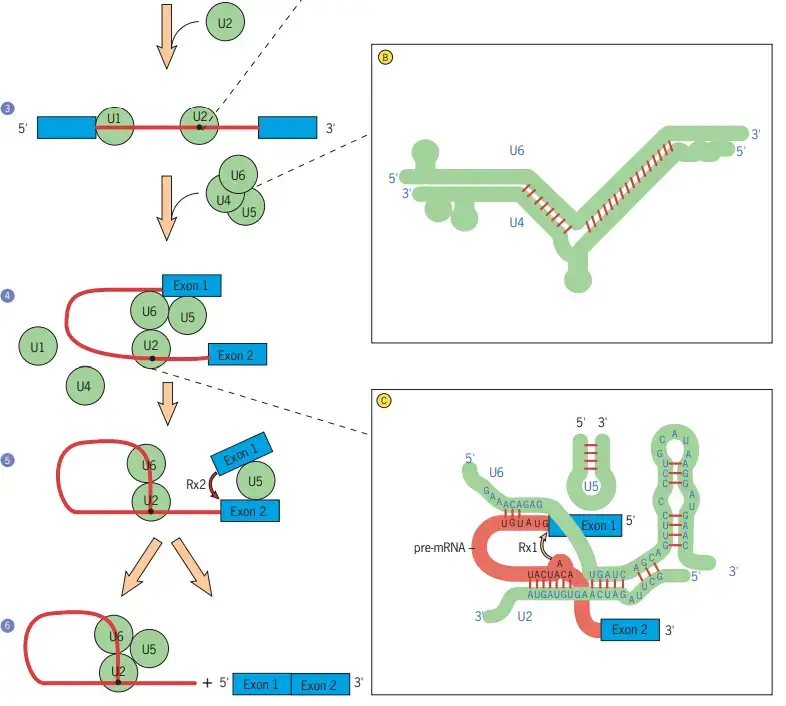
- The subsequent stage is the binding of the U4/U6 and U5 snRNPs to the premRNA, followed by the displacement of the U1 snRNP (step 4).
- A spliceosome is assembled through a series of dynamic interactions between pre-mRNA and specific snRNAs, as well as among the snRNAs themselves.
- As they join the pre-mRNA complex, the U4 and U6 snRNAs base-pair extensively with one another (inset B).
- The U4 snRNA is then removed, and the portions of U6 that were base-paired with U4 become base-paired with a piece of the U2 snRNA (inset C).
- A piece of the U6 snRNA is present at the 5 splice site (inset C), displacing the U1 snRNA that was bound there previously (inset A). It is hypothesised that U6 is a ribozyme and U4 inhibits its catalytic activity.
- Once the U1 and U4 snRNAs have been displaced, the U6 snRNA is in a position to catalyse the two chemical processes required for intron elimination, according to this hypothesis.
- According to a different theory, the processes are catalysed by the joint activity of U6 snRNA and a U5 snRNP protein. Regardless of the mechanism, the first reaction (shown by the arrow in inset C) cleaves the 5 splice site, producing a free 5 exon and a lariat intron–3 exon intermediate (step 5).
- It is believed that the free 5 exon is maintained in place by the U5 snRNA of the spliceosome, which also interacts with the 3 exon (step 5).
- A second cleavage reaction at the 3 splice site excises the lariat intron and simultaneously connects the ends of the two adjacent exons (arrow, step 5). (step 6).
- After splicing, the snRNPs must be freed from the pre-mRNA, the original relationships between snRNAs must be restored, and the snRNPs must be rebuilt at other intron sites.
What is tRNA splicing?
- tRNA (also known as tRNA-like) splicing is an uncommon type of splicing that often happens in tRNA.
- The biology of the splicing reaction is distinct from that of the spliceosomal and self-splicing pathways.
- In Saccharomyces cerevisiae, a yeast tRNA splicing endonuclease heterotetramer consisting of TSEN54, TSEN2, TSEN34, and TSEN15 cleaves pre-tRNA at two places in the acceptor loop to generate a 5′-half tRNA terminating at a 2′,3′-cyclic phosphodiester group and a 3′-half tRNA terminating at a 5′
- The 5′-hydroxyl group is then phosphorylated by yeast tRNA kinase utilising adenosine triphosphate.
- The cyclic phosphodiester group is cleaved by yeast tRNA cyclic phosphodiesterase to create a 2′-phosphorylated 3′ end.
- Yeast tRNA ligase connects the two halves by adding an adenosine monophosphate group to the 5′ end of the 3′-half.
- 2′-phosphotransferase dependent on NAD then removes the 2′-phosphate group.
RNA Splicing errors
- RNA splicing errors can have significant implications for gene expression and protein function. The splicing of nuclear pre-mRNAs is a crucial process in metazoan organisms, but mutations can lead to errors in splicing, giving rise to various splicing-related diseases. These errors primarily occur in alternative splicing, resulting in the production of non-functional biological products.
- One common type of error is caused by mutations at the splice site. These mutations can lead to the loss of exons or the inclusion of an intron, disrupting the normal functioning of the RNA sequence. Such disruptions can have severe consequences for gene expression and protein synthesis.
- Another type of error arises from the displacement of a splice site. When a splice site is displaced, it can lead to the formation of longer or shorter exons than intended. As a result, erroneous products may be generated, further compromising the functionality of the RNA molecule.
- In organisms like plants, stress-induced alternative splicing plays a significant role in various metabolic pathways. However, this alternative splicing can introduce changes in the normal functioning of the plant due to errors in splicing. The misregulation of alternative splicing under stressful conditions can impact gene expression patterns and disrupt metabolic processes.
- Eukaryotic cells with a high degree of alternative splicing are more susceptible to splicing errors. This is because the splice sites in these cells have evolved to offer a weak binding potential for components of the spliceosome, making accurate splice-site pairing more challenging.
- Furthermore, the accuracy of splice-site pairing is also influenced by the accuracy of transcription. Transcription machinery occasionally makes mistakes during the nucleotide insertion step, occurring at a rate of approximately once in every 103–105 nucleotides. These transcription errors can further contribute to splicing errors and affect the overall fidelity of gene expression.
- In conclusion, errors in RNA splicing can arise from various sources, including mutations at splice sites, displacement of splice sites, stress-induced alternative splicing, and transcription errors. These errors can have profound consequences for gene expression, protein functionality, and overall cellular processes. Understanding and addressing these splicing errors are essential for elucidating the molecular basis of splicing-related diseases and developing potential therapeutic interventions.
Application/Importance of RNA Splicing
RNA splicing mistakes can have serious consequences in a variety of biological and medicinal applications. Here are some major aspects about the significance of pre-mature RNA splicing and its relationship to several areas:
- Protein variety: Pre-mRNA splicing is a critical step in cellular metabolism that contributes to protein variety. Changes in the number and sequence of exons and introns in the RNA sequence cause this diversity. Cells can create several protein isoforms from a single gene by selectively eliminating or adding certain exons during splicing, hence increasing the functional repertoire of the proteome.
- Gene and Protein Regulation: RNA splicing is essential for controlling gene and protein composition within cells. Cells can fine-tune gene expression and control the creation of specific protein variants by modifying splicing patterns. Cells can respond to developmental signals, environmental changes, and physiological demands thanks to this control, which ensures appropriate cellular function and homeostasis.
- Protein Evolution: RNA splicing contributes to the evolution of new and better proteins. Organisms can develop unique protein isoforms with distinct functions and characteristics by using alternative splicing. This process allows organisms to respond to evolutionary stresses and optimize their biological processes by providing a mechanism for adaptation and diversity.
- Cancer Markers and Therapy Targets: Aberrant splicing isoforms have been discovered as cancer markers and prospective cancer therapy targets. Splicing events that are misregulated can result in the generation of aberrant protein isoforms that contribute to the genesis and progression of cancer. Specific splicing variations linked to cancer may offer therapeutic prospects for intervention and treatment techniques.
- Cancer Pathology: Pre-mRNA splicing is important in cancer pathology, particularly in regulating three functional aspects: proliferation, metastasis, and apoptosis. Splicing events that are dysregulated can change the expression of genes involved in cell proliferation, contribute to the acquisition of metastatic capabilities, and alter the balance between cell survival and cell death. Understanding and modifying cancer-related splicing patterns might provide vital insights into disease mechanisms and potentially open up new treatment approaches.
FAQ
What is RNA splicing?
RNA splicing is a process in which non-coding introns are removed from pre-messenger RNA (pre-mRNA) molecules, and the remaining coding exons are joined together to form a mature mRNA molecule.
Why is RNA splicing important?
RNA splicing is essential for generating protein diversity. It allows multiple protein isoforms to be produced from a single gene by selectively including or excluding different exons during the splicing process.
What are splice sites?
Splice sites are specific sequences at the boundaries between exons and introns. They contain consensus sequences that are recognized by the spliceosome, a complex of proteins and small nuclear RNAs (snRNAs) involved in the splicing process.
What are alternative splicing and its significance?
Alternative splicing refers to the process by which different combinations of exons are included or excluded from the final mRNA transcript. It greatly expands the coding potential of the genome and allows for the production of multiple protein isoforms with different functions from a single gene.
Can errors occur during RNA splicing?
Yes, errors can occur during RNA splicing. Mutations or variations in the splice sites or other splicing regulatory elements can lead to incorrect splicing patterns, resulting in the production of abnormal mRNA isoforms or non-functional proteins.
What are splicing factors?
Splicing factors are proteins that bind to pre-mRNA molecules and regulate the splicing process. They help determine which exons are included or excluded during splicing by interacting with the splice sites and other splicing regulatory elements.
How is RNA splicing related to genetic diseases?
Mutations in splice sites, splicing factors, or other splicing regulatory elements can disrupt normal splicing patterns and lead to genetic diseases. These diseases are often characterized by abnormal protein function due to errors in RNA splicing.
Can RNA splicing be influenced by environmental factors?
Yes, environmental factors and cellular conditions can influence RNA splicing. Stress, disease, or changes in cellular signaling pathways can alter splicing patterns, leading to the production of different protein isoforms and potentially affecting cellular function.
Are there any drugs or therapies targeting RNA splicing?
Yes, there is increasing interest in developing therapeutics that target RNA splicing. Some drugs aim to correct splicing errors or modify splicing patterns in diseases caused by splicing defects. These approaches hold promise for treating certain genetic disorders and cancers.
Can RNA splicing be studied experimentally?
Yes, researchers use various experimental techniques to study RNA splicing. These include RNA sequencing (RNA-seq) to analyze splicing patterns, genetic engineering to manipulate splice sites or splicing factors, and biochemical assays to investigate the interactions between splicing components.
References
- Jurica, M. S., & Roybal, G. A. (2013). RNA Splicing. Encyclopedia of Biological Chemistry, 185–190. doi:10.1016/b978-0-12-378630-2.00674-5
- Blumenthal, T. (2013). Trans-Splicing. Brenner’s Encyclopedia of Genetics, 170–172. doi:10.1016/b978-0-12-374984-0.01575-8
- Hsu, S.-N., & Hertel, K. J. (2009). Spliceosomes walk the line: Splicing errors and their impact on cellular function. RNA Biology, 6(5), 526–530. doi:10.4161/rna.6.5.9860
- WANG, Y., LIU, J., HUANG, B., XU, Y.-M., LI, J., HUANG, L.-F., … WANG, X.-Z. (2014). Mechanism of alternative splicing and its regulation. Biomedical Reports, 3(2), 152–158. doi:10.3892/br.2014.407
- Winter AJ, Groot Koerkamp MJ, Tabak HF. The mechanism of group I self-splicing: an internal guide sequence can be provided in trans. EMBO J. 1990 Jun;9(6):1923-8. doi: 10.1002/j.1460-2075.1990.tb08319.x. PMID: 2189725; PMCID: PMC551900.
- Van den Hoogenhof, M. M. G., Pinto, Y. M., & Creemers, E. E. (2016). RNA Splicing. Circulation Research, 118(3), 454–468. doi:10.1161/circresaha.115.307872
- https://en.wikipedia.org/wiki/Exon
- https://www.technologynetworks.com/genomics/articles/alternative-splicing-importance-and-definition-351813
- https://en.wikipedia.org/wiki/RNA_splicing
- https://www.biointeractive.org/classroom-resources/rna-splicing
- https://openstax.org/books/biology-2e/pages/15-4-rna-processing-in-eukaryotes
- https://www.slideshare.net/msaltyy/rna-splicing-65637051
- https://www.slideshare.net/VaishaliC4/rna-splicing-250748216
- https://www.slideshare.net/aabuans/spliceosome-250916824
- http://web.mit.edu/hst.160/www/quiz/Splicing.htm
- https://www.nature.com/scitable/topicpage/rna-splicing-introns-exons-and-spliceosome-12375/
- https://microbenotes.com/rna-splicing/