What is RNA Polymerase?
- RNA polymerase is an essential enzyme involved in the transcription process, where it catalyzes the synthesis of RNA molecules from a DNA template. It plays a critical role in the transfer of genetic information from DNA to RNA.
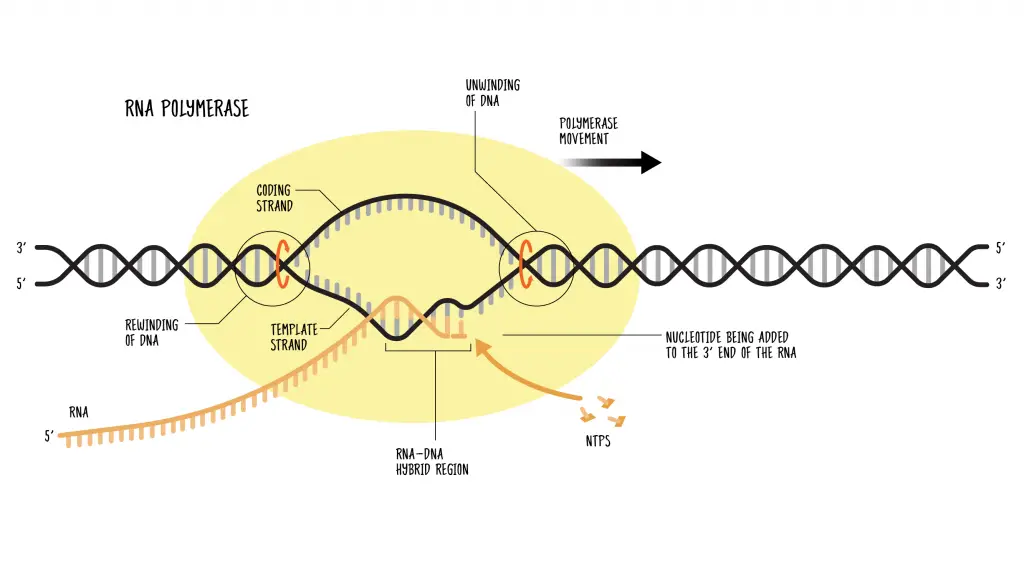
- During transcription, RNA polymerase binds to a specific region on the DNA called the promoter. This interaction triggers the unwinding of the DNA double helix, allowing the enzyme to access the template strand. The RNA polymerase then begins synthesizing an RNA molecule that is complementary to the DNA template.
- The process of RNA synthesis occurs in the 5′ to 3′ direction, with the RNA polymerase moving along the DNA template strand in the 3′ to 5′ direction. As it progresses, the enzyme adds nucleotides to the growing RNA strand, based on the complementary base pairing with the DNA template.
- RNA polymerase functions in collaboration with various proteins to carry out transcription. These proteins assist in the specific binding of RNA polymerase to the DNA, facilitate the unwinding of the DNA strands, modulate the enzymatic activity of RNA polymerase, and regulate the speed and fidelity of transcription.
- Different organisms have varying types and compositions of RNA polymerases. Bacteria typically have a single type of RNA polymerase, while eukaryotes, such as multicellular organisms and yeasts, possess three distinct types. Despite these differences, the general transcriptional mechanisms share similarities across species.
- Regulation of transcription is crucial for achieving spatial and temporal changes in gene expression. Various mechanisms control the activity of RNA polymerase, allowing genes to be selectively transcribed in response to different cellular signals and developmental stages.
- In conclusion, RNA polymerase is an enzyme responsible for the synthesis of RNA molecules from a DNA template during transcription. It plays a vital role in the transfer of genetic information and is involved in complex interactions with other proteins to ensure accurate and controlled gene expression.
Definition RNA Polymerase
RNA polymerase is an enzyme that catalyzes the synthesis of RNA molecules using a DNA template during the process of transcription.
Features of RNA Polymerase
RNA polymerase is a versatile enzyme with several key features that contribute to its essential role in transcription. Here are some prominent features of RNA polymerase:
- Template-dependent synthesis: RNA polymerase catalyzes the synthesis of RNA molecules using a DNA template. It reads the DNA template in the 3′ to 5′ direction and synthesizes the RNA molecule in the complementary 5′ to 3′ direction.
- Promoter recognition: RNA polymerase recognizes specific DNA sequences called promoters, which mark the starting points for transcription. Promoters contain consensus sequences that are recognized by RNA polymerase and its associated factors.
- DNA unwinding: Once bound to the promoter, RNA polymerase uses helicase activity to locally unwind the double-stranded DNA. This unwinding allows access to the DNA template strand for RNA synthesis.
- Initiation and elongation: After DNA unwinding, RNA polymerase initiates RNA synthesis by adding the first nucleotide to the growing RNA chain. It continues to elongate the RNA molecule by adding subsequent nucleotides in a stepwise manner, guided by complementary base pairing with the DNA template.
- Proofreading and editing: RNA polymerase possesses intrinsic proofreading capabilities. It can recognize and remove incorrectly added nucleotides during RNA synthesis through a mechanism known as backtracking. This proofreading activity helps maintain the accuracy of the transcribed RNA sequence.
- Termination recognition: RNA polymerase recognizes specific termination signals in the DNA sequence that mark the end of transcription. These signals cause the RNA polymerase to dissociate from the DNA template and release the synthesized RNA molecule.
- Transcription factors and co-factors: RNA polymerase often interacts with various transcription factors and co-factors to regulate its activity and enhance transcription efficiency. These factors can modulate the binding of RNA polymerase to the promoter, influence the rate of transcription, and coordinate gene expression.
- Diversity of RNA polymerases: Different types of RNA polymerases exist in cells, each with distinct roles. For example, RNA polymerase I is responsible for synthesizing ribosomal RNA (rRNA), RNA polymerase II transcribes protein-coding genes, and RNA polymerase III synthesizes transfer RNA (tRNA) and other small non-coding RNAs.
- Regulation of transcription: Transcription can be regulated through the interaction of RNA polymerase with regulatory proteins and DNA sequences. Factors such as enhancers, silencers, and transcriptional activators or repressors can influence RNA polymerase binding and activity, allowing precise control of gene expression.
- Evolutionary conservation: RNA polymerase is present in all domains of life, including bacteria, archaea, and eukaryotes, underscoring its fundamental importance in cellular processes. While there are differences in the subunit composition and regulation of RNA polymerase across different organisms, the core enzymatic mechanism is conserved.
Structure of RNA Polymerase
The structure of RNA polymerase (RNAP) is essential for its function in transcription, where it synthesizes RNA molecules from DNA templates. Here are the key features of the RNA polymerase structure:
- Core RNA Polymerase: In prokaryotes, such as Escherichia coli (E. coli), the core RNA polymerase consists of five subunits. It includes two alpha (α) subunits, each weighing around 36 kDa, a beta (β) subunit weighing approximately 150 kDa, a beta prime (β’) subunit weighing about 155 kDa, and a small omega (ω) subunit. The core RNA polymerase is responsible for catalyzing the synthesis of RNA.
- Holoenzyme Formation: The core RNA polymerase associates with a sigma (σ) factor to form a holoenzyme. The sigma factor plays a crucial role in the recognition and binding of specific DNA sequences called promoters. The binding of the sigma factor to the core enzyme forms the active holoenzyme, allowing transcription initiation at specific promoter sites. Once transcription begins, the sigma factor may dissociate, and the core enzyme continues with the elongation phase.
- Structural Configuration: The core RNA polymerase complex exhibits a distinctive structure often referred to as a “crab claw” or “clamp-jaw” due to its shape. This structure includes an internal channel that runs along the length of the enzyme, providing a pathway for the DNA template strand and newly synthesized RNA strand during transcription. The channel allows the movement of nucleotides for RNA synthesis.
- Additional Subunits: Eukaryotic and archaeal RNA polymerases have a more complex structure compared to prokaryotic RNA polymerases. They consist of additional subunits that contribute to their functionality and regulation. These extra subunits provide stability, enhance the binding affinity to DNA, and interact with other transcription factors and regulators.
- Metal Cofactors: RNA polymerases, including both prokaryotic and eukaryotic forms, contain metal cofactors that facilitate their enzymatic activity during transcription. Zinc and magnesium cations are particularly important metal cofactors that assist in various steps of the transcription process. They help stabilize the structure of the RNA polymerase and participate in catalytic reactions.
The detailed understanding of the RNA polymerase structure, as elucidated by studies like Roger D. Kornberg’s work, has provided valuable insights into the mechanisms of transcription and the regulation of gene expression. The diverse subunits and metal cofactors contribute to the efficiency, accuracy, and regulatory control of RNA synthesis by RNA polymerase.
Types of RNA polymerase
Prokaryotic (Bacteria, archaea, viruses) organisms possess one kind of RNA polymerase which synthesizes all subtypes the RNA. Eukaryotes (multicellular organisms) contain five different kinds of RNA polymerases that perform different roles in the synthesis of various RNA molecules.
Prokaryotic RNA polymerase
Prokaryotic RNA polymerase is a single type of RNA polymerase found in prokaryotes, and it plays a central role in the transcription of all classes of RNA, including mRNA, tRNA, rRNA, and sRNA.
The RNA polymerase molecule consists of two main domains: the core and the holoenzyme. It is composed of five subunits: β, β’, α (αI and αII), and ω.
The β and β’ subunits together form the catalytic center of the RNA polymerase, responsible for the synthesis of RNA molecules.
The α subunit is composed of two distinct domains: the N-terminal domain (α-NTD) and the C-terminal domain. The N-terminal domain is involved in dimerization, forming a2 and facilitating the assembly of the RNA polymerase. The C-terminal domain plays a role in binding to the Upstream Promoter (UP) DNA sequence at promoters for rRNA and tRNA genes and interacts with various transcriptional activators.
Each subunit of prokaryotic RNA polymerase has specific functions:
- β subunit: It has a size of 150.4 kDa and, together with the β’ subunit, forms the catalytic center responsible for RNA synthesis.
- β’ subunit: With a size of 155.0 kDa, it also contributes to the catalytic center and participates in RNA synthesis.
- α subunit (αI and αII): With a size of 36.5 kDa, the α subunit is involved in the assembly of the RNA polymerase complex. It also binds to the UP sequence in the promoter region.
- ω subunit: It has a size of 155.0 kDa and confers specificity for promoters. The ω subunit binds to the -10 and -35 sites in the promoter region.
The promoter region is a specific DNA sequence that is essential for the accurate and specific initiation of transcription. It is at the promoter that the RNA polymerase binds accurately to initiate the transcription process.
The prokaryotic RNA polymerase, with its subunit composition and distinct domains, allows for the efficient synthesis of different classes of RNA in prokaryotic organisms. Its specificity for promoters and interactions with other regulatory factors ensure accurate transcription initiation and gene expression.
| Subunit | Size | Function |
| β | 150.4 kDa | The β’ + β form the catalytic center, responsible for RNA synthesis. |
| β’ | 155.0 kDa | The β’ + β form the catalytic center, responsible for RNA synthesis. |
| α (αI and αII) | 36.5 kDa | It is made up of the enzyme assembly, and it also binds the UP sequence in the promoter. |
| ω | 155.0 kDa | It confers specificity for promoter; and binds to -10 and -35 sites in the promoter. |
Eukaryotic RNA polymerase
Eukaryotic RNA polymerase is a group of enzymes responsible for synthesizing different types of RNA molecules in eukaryotic organisms. There are five known types of eukaryotic RNA polymerases, each with specific functions:
- RNA polymerase I: Located in the nucleolus, RNA polymerase I synthesizes a precursor of ribosomal RNA (pre-rRNA) called 45S (or 35S in yeast). This pre-rRNA matures and forms the major RNA components of ribosomes, which are essential for protein synthesis.
- RNA polymerase II: Found in the nucleus, RNA polymerase II is a complex enzyme consisting of approximately 12 subunits. It is responsible for transcribing the precursors of messenger RNA (pre-mRNA), which undergo further processing to become mature mRNA molecules. RNA polymerase II plays a crucial role in the transcription of most eukaryotic genes.
- RNA polymerase III: Also located in the nucleus, RNA polymerase III transcribes different types of small RNA molecules, including transfer RNA (tRNA), 5S ribosomal RNA (rRNA), and other small non-coding RNAs. These molecules are involved in various cellular processes and are essential for normal cellular functioning.
- RNA polymerase IV and V: These two RNA polymerases are unique to plants. Their precise functions are not yet fully understood, but they are involved in the synthesis of small interfering RNA (siRNA) and the formation of heterochromatin in the cell nucleus. RNA polymerase IV and V are primarily found in the nucleus and exhibit similarities to bacterial RNA polymerases.
Eukaryotic RNA polymerases are large protein complexes with multiple subunits. They range from 8 to 14 subunits, and their molecular weights can be approximately 500,000 for each polymerase. The subunits, such as α, β, and β’, play critical roles in catalysis and protein assembly.
Each RNA polymerase in eukaryotes has a specific cellular localization and distinct targets for transcription. They are involved in transcribing different types of RNA molecules necessary for various cellular processes, including protein synthesis, ribosome assembly, and regulation of gene expression.
Eukaryotic cells possess multiple types of RNA polymerases (RNAPs) located in the nucleus, each responsible for the synthesis of specific subsets of RNA molecules. Despite their functional differences, all eukaryotic RNAPs share structural and mechanistic similarities with each other and with bacterial RNAP.
RNA polymerase I (RNAP I) is responsible for synthesizing a precursor molecule called pre-rRNA (45S in most eukaryotes and 35S in yeast). This pre-rRNA undergoes maturation to produce the major components of the ribosome, namely the 28S, 18S, and 5.8S rRNAs. These rRNAs play crucial roles in the translation process.
RNA polymerase II (RNAP II) is the most extensively studied type of eukaryotic RNAP. It synthesizes precursor molecules for messenger RNA (mRNA), as well as most small nuclear RNAs (snRNAs) and microRNAs. Transcription by RNAP II requires the involvement of various transcription factors that facilitate its binding to specific promoter regions on DNA. This precise control over transcription allows for the regulation of gene expression.
RNA polymerase III (RNAP III) is responsible for synthesizing transfer RNAs (tRNAs), 5S rRNA, and other small nuclear and cytoplasmic RNAs. These molecules play essential roles in protein synthesis and other cellular processes.
RNA polymerase IV (RNAP IV) and RNA polymerase V (RNAP V) are unique to plants. RNAP IV is involved in the synthesis of small interfering RNAs (siRNAs), which play a role in gene silencing and defense against viruses. RNAP V is responsible for synthesizing RNAs involved in siRNA-directed heterochromatin formation, contributing to gene regulation in plants.
Within eukaryotic cells, chloroplasts possess their own RNAPs, known as plastid-encoded polymerases (PEP). These RNAPs are highly similar to bacterial RNAPs and utilize sigma factors encoded in the nuclear genome to initiate transcription. Additionally, eukaryotic chloroplasts contain another type of RNAP called nucleus-encoded polymerase (NEP), which is structurally and mechanistically unrelated to PEP.
Eukaryotic mitochondria also possess their own RNAP, known as POLRMT in humans. This nucleus-encoded single-subunit RNAP is functionally similar to phage-like polymerases and plays a vital role in mitochondrial gene expression.
The presence of multiple types of eukaryotic RNAPs reflects the complexity and diversity of RNA synthesis and regulation in these organisms. Understanding the functions and mechanisms of these RNAPs provides insights into gene expression, cellular processes, and the overall functioning of eukaryotic cells.
Functions of RNA Polymerase
RNA polymerase (RNAP) is a crucial enzyme involved in the process of gene transcription, where it synthesizes RNA molecules from DNA templates. Here are the main functions of RNA polymerase:
- Initiation of Transcription: RNA polymerase recognizes and binds to specific DNA sequences called promoters, which mark the starting points for transcription. It initiates the synthesis of an RNA chain by adding the first nucleotide to the growing RNA strand.
- Elongation of RNA Strand: Once transcription is initiated, RNA polymerase moves along the DNA template strand and synthesizes an RNA molecule that is complementary to the DNA sequence. This process is called elongation. RNA polymerase can add nucleotides to the growing RNA chain, extending it in a 5′ to 3′ direction.
- Termination of Transcription: RNA polymerase recognizes specific DNA sequences at the end of genes, known as terminators. These sequences signal the termination of transcription, and RNA polymerase releases the newly synthesized RNA molecule and detaches from the DNA template.
- Production of mRNA: One of the major products of RNA polymerase is messenger RNA (mRNA). mRNA carries the genetic information from DNA to the ribosomes, where it serves as a template for protein synthesis during translation.
- Non-coding RNA Genes: RNA polymerase transcribes various non-coding RNA genes, which produce functional RNA molecules that are not translated into proteins. Examples include transfer RNA (tRNA) that helps incorporate specific amino acids into growing polypeptide chains during translation, and ribosomal RNA (rRNA) that forms a structural component of ribosomes involved in protein synthesis. Other non-coding RNA genes, such as microRNA, play important roles in regulating gene activity.
- Catalytic RNA: RNA polymerase can also transcribe catalytic RNA molecules known as ribozymes. These RNA molecules exhibit enzymatic activity and can catalyze specific biochemical reactions.
RNA polymerase vs DNA polymerase
DNA polymerase creates double-stranded molecules by separating DNA strands from their unwound form when it is replicating. Although the final products of transcription and replication are distinct, they each work on DNA by introducing nucleotides in the same 5′-3 direction. Contrary to RNA polymerase, DNA Polymerase is semi-conserved process that makes use of both strands of the double-stranded DNA molecule to serve as the template to replicate.
| Comparison | RNA Polymerase | DNA Polymerase |
| Function | Transcription of DNA | DNA replication |
| Scope | To create DNA copies of genes | To duplicate the whole genome |
| The time of the event | It is used in transcription in transcription G phase(s) | In replication, it is utilized during S phase |
| Primer | Not necessary to be transcribed | Required to initiate replication |
| Base pairs are the components used to synthesize products | Adenine, Guanine, Cytosine and Uracil | Adenine, Guanine, Cytosine and Thymine |
| The product that results | Single-strandedRNAs (e.g. mRNA) | Double-stranded DNA |
RNA polymerase in Archaea
- Archaea, one of the three domains of life, possess a single type of RNA polymerase (RNAP) responsible for transcribing all types of RNA molecules. The structure and mechanism of archaeal RNA polymerase closely resemble those of bacterial RNA polymerase and eukaryotic nuclear RNA polymerases I-V. In particular, archaeal RNA polymerase shares significant structural and mechanistic similarities with eukaryotic nuclear RNA polymerase II.
- The discovery of archaeal RNA polymerase is relatively recent. The first analysis of an archaeal RNAP was conducted in 1971, focusing on the RNAP of the extreme halophile Halobacterium cutirubrum. Crystal structures of RNA polymerases from Sulfolobus solfataricus and Sulfolobus shibatae have provided insights into the architecture of archaeal RNA polymerase. These studies have identified a total of thirteen subunits in archaeal RNA polymerase.
- In archaea, the subunit corresponding to the eukaryotic Rpb1 (the largest subunit of RNA polymerase II) is split into two subunits. However, there is no homolog to the eukaryotic Rpb9 subunit (POLR2I) in the Sulfolobus shibatae complex. Instead, a subunit called TFS (transcription factor S), which bears similarity to TFIIS, has been proposed as a potential functional equivalent. Another additional subunit, referred to as Rpo13, is present in archaeal RNA polymerase. Rpo13, along with Rpo5, occupies a space that corresponds to an insertion found in bacterial β’ subunits. Notably, Rpo3 in archaeal RNA polymerase is an iron-sulfur protein, distinguishing it from its counterparts in other domains of life. There is also a shared sequence similarity between the archaeal RNAP I/III subunit AC40 and certain eukaryotic RNAP subunits, although the iron-binding function is absent in AC40.
- In the past, archaeal RNAP subunits were designated using an “RpoX” nomenclature, where each subunit was assigned a letter independent of other systems. However, in 2009, a new nomenclature was proposed based on the numbering system of eukaryotic RNA polymerase II subunits, using the “Rpb” prefix.
- The study of archaeal RNA polymerase has provided valuable insights into the transcription process in archaea and has deepened our understanding of the structural and functional relationships between RNA polymerases across different domains of life.
RNA polymerase in Viruses
- Viruses employ various strategies for RNA synthesis, and their RNA polymerases (RNAPs) exhibit diverse structures and mechanisms. Orthopoxviruses and certain other nucleocytoplasmic large DNA viruses encode their own multi-subunit RNAPs, which bear similarity to eukaryotic RNAPs. However, these viral RNAPs often possess minified or missing subunits compared to their eukaryotic counterparts. The specific eukaryotic RNAP to which they are most similar is a subject of debate within the scientific community. On the other hand, many other viruses utilize single-subunit DNA-dependent RNAPs (ssRNAPs) for RNA synthesis. These ssRNAPs share structural and mechanistic similarities with the single-subunit RNAPs found in eukaryotic chloroplasts (RpoT) and mitochondria (POLRMT). Examples of widely studied single-subunit RNAPs include the bacteriophage T7 RNA polymerase. Notably, ssRNAPs lack proofreading capabilities.
- In the case of B. subtilis prophage SPβ, a distinct monomeric RNAP is formed using YonO, a homolog of the β and β’ subunits found in multi-subunit RNAPs. This unique RNAP structure diverged significantly from the canonical five-unit multi-subunit RNAP, potentially predating the last universal common ancestor.
- In addition to DNA-dependent RNAPs, some viruses employ RNA-dependent RNAPs, which utilize RNA as a template for RNA synthesis instead of DNA. This mechanism is observed in negative strand RNA viruses and dsRNA viruses, which spend a portion of their life cycle as double-stranded RNA. Interestingly, certain positive strand RNA viruses, such as poliovirus, also possess RNA-dependent RNAPs.
- The diversity of RNA polymerases in viruses reflects their adaptation to different genetic and replicative strategies. The study of viral RNA polymerases enhances our understanding of viral replication mechanisms and provides potential targets for antiviral interventions.
FAQ
What is RNA polymerase?
RNA polymerase is an enzyme responsible for synthesizing RNA molecules from a DNA template during the process of transcription. It plays a crucial role in gene expression by catalyzing the formation of RNA chains that are complementary to the DNA sequence.
How many types of RNA polymerase are there?
There are multiple types of RNA polymerase in different organisms. In eukaryotes, there are three main types: RNA polymerase I, RNA polymerase II, and RNA polymerase III. Additionally, plants have RNA polymerase IV and RNA polymerase V, and bacteria have a single type of RNA polymerase.
What are the functions of different RNA polymerases?
Each type of RNA polymerase has specific functions. RNA polymerase I synthesizes ribosomal RNA (rRNA), RNA polymerase II transcribes messenger RNA (mRNA) and several small RNAs, and RNA polymerase III is responsible for producing transfer RNA (tRNA), 5S rRNA, and other small nuclear and cytoplasmic RNAs.
How does RNA polymerase recognize the starting point for transcription?
RNA polymerase recognizes specific DNA sequences called promoters, which indicate the starting point for transcription. Promoters contain specific nucleotide sequences that serve as binding sites for transcription factors, allowing RNA polymerase to initiate transcription at the correct location.
How does RNA polymerase unwind the DNA during transcription?
RNA polymerase has helicase activity, meaning it can unwind the DNA double helix as it progresses along the DNA template. This unwinding allows the enzyme to access the DNA strand for RNA synthesis.
What is the role of sigma factors in RNA polymerase function?
In bacteria, sigma factors play a crucial role in guiding RNA polymerase to specific promoter sequences on DNA. Sigma factors help RNA polymerase recognize and bind to the correct promoter regions, ensuring accurate initiation of transcription.
Can RNA polymerase proofread and correct errors in RNA synthesis?
RNA polymerase lacks proofreading activity, unlike DNA polymerase. As a result, it can introduce errors during RNA synthesis. However, the cell has other mechanisms, such as RNA editing and degradation pathways, to correct or eliminate faulty RNA molecules.
Are there any other types of RNA polymerase in cellular organelles?
Yes, eukaryotic cells possess RNA polymerases in their cellular organelles. For example, chloroplasts have plastid-encoded polymerases (PEP) involved in chloroplast gene expression. Mitochondria have their own RNA polymerase called POLRMT, which transcribes mitochondrial genes.
Are there any viral RNA polymerases?
Yes, viruses also possess RNA polymerases for their replication and gene expression. Some viruses have their own RNA polymerases, while others can utilize the host cell’s RNA polymerase machinery for transcription.
How is RNA polymerase regulated?
RNA polymerase activity is tightly regulated to ensure proper gene expression. Transcription factors, coactivators, and repressors bind to specific DNA sequences or interact with RNA polymerase to control its activity. Additionally, various signaling pathways and environmental factors can influence the activity of RNA polymerase and gene transcription.
References
- Cramer, P. (2019). Organization and regulation of gene transcription. Nature, 573(7772), 45-54.
- Ebright, R. H., & Ebright, Y. W. (2017). Transcription activation by structurally diverse activators: the benefits of promiscuity. Trends in biochemical sciences, 42(7), 502-513.
- Fazal, F. M., Meng, C. A., Murakami, K. S., & Kornberg, R. D. (2017). Block of RNA polymerase II transcription by stalled RNA polymerase I. Science, 358(6366), 1283-1291.
- Nudler, E. (2012). RNA polymerase backtracking in gene regulation and genome instability. Cell, 149(7), 1438-1445.
- Svetlov, V., & Nudler, E. (2013). Basic mechanism of transcription by RNA polymerase II. Biochimica et Biophysica Acta (BBA)-Gene Regulatory Mechanisms, 1829(1), 20-28.
- Werner, F., & Grohmann, D. (2011). Evolution of multisubunit RNA polymerases in the three domains of life. Nature Reviews Microbiology, 9(2), 85-98.
- Zhang, G., Campbell, E. A., Minakhin, L., Richter, C., Severinov, K., & Darst, S. A. (1999). Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell, 98(6), 811-824.
- Zhang, J., Landick, R., & Gelles, J. (2019). Mechanisms of bacterial transcription termination: All good things must end. Annual Review of Biochemistry, 88, 319-347.