What is Reverse Transcription PCR (RT-PCR)?
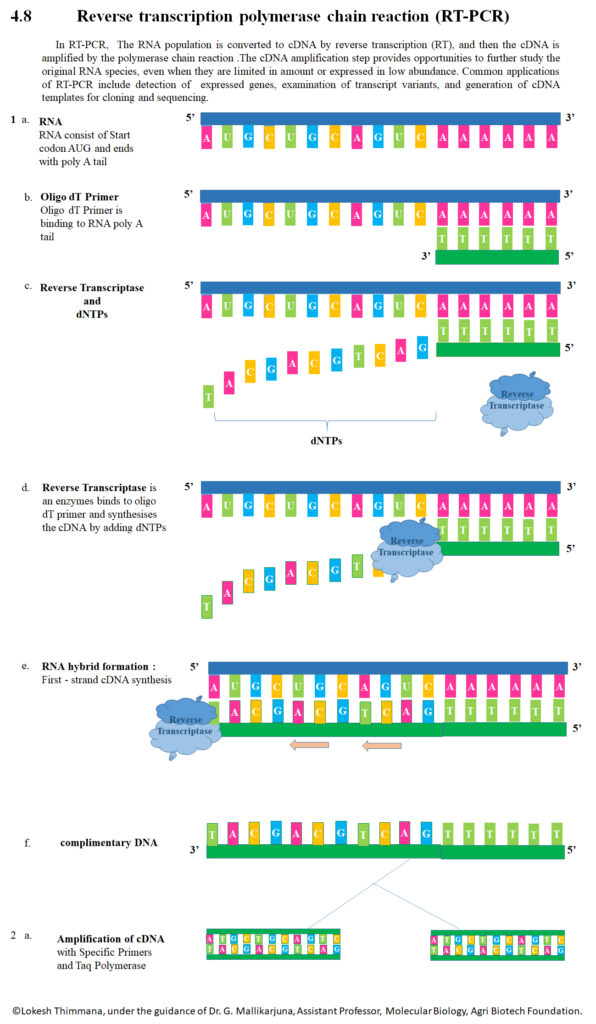
Reverse Transcription Polymerase Chain Reaction (RT-PCR) is defined as a lab method in which RNA is converted into complementary DNA (cDNA) by an enzyme called reverse transcriptase, then that cDNA is amplified via PCR.
The technique is used for detection, quantification of specific RNA molecules (eg mRNA, viral RNA) in samples.
The process is begun with RNA isolation from cells, tissues, or bodily fluids, which is essential because quality of RNA strongly affects the outcome.
After isolation, reverse transcription is performed: reverse transcriptase enzyme + primers (which may be oligo-dT, random primers, or gene-specific) are used so that cDNA is synthesised from RNA template.
PCR amplification is performed on the cDNA, using DNA polymerase and specific primers; cycles of denaturation, annealing, extension are repeated to exponentially increase amount of target DNA.
The detection of the amplified product may be done either at end-point (after PCR cycles, eg by gel electrophoresis) or in real-time (quantitative RT-PCR, using fluorescent probes/dyes) so that quantification is possible.
The sensitivity of RT-PCR is high, very small amounts of RNA can be detected; method is more sensitive than e.g. Northern blot or RNase protection assay.
Limitations are present: RNA degradation can lead to false / misleading results, efficiency of reverse transcription may vary, risk of contamination especially when multiple handling steps are involved.
Variants of RT-PCR are used: one-step RT-PCR (reverse transcription + PCR in same tube) and two-step RT-PCR (RT first, then PCR separately) are common, each has trade-offs in flexibility, contamination risk etc.12345
Objectives of Reverse Transcription PCR (RT-PCR)
- Amplification of Specific RNA Sequences is aimed, so that a small amount of RNA can be made into large number of copies, enabling detection of target transcripts.
- Detection of Gene Expression Levels is enabled, in order that changes in expression under different conditions / treatments may be studied.
- Quantification of RNA Amounts is permitted, so that relative or absolute amounts of messenger RNA (mRNA) or viral RNA may be measured.
- Diagnostics of Infectious Diseases is facilitated, because RNA viruses etc can be detected in clinical samples by RT-PCR.
- Early Detection is achieved, since even very low copy‐numbers of RNA can be amplified and detected, allowing diagnosis before obvious symptoms or high pathogen loads.
- Gene Transcript Profiling is supported, allowing many transcripts from a sample to be compared, for e.g. in developmental biology, stress responses, or disease states.678
Principle of Reverse Transcription PCR (RT-PCR)
The Principle is based on conversion of RNA into complementary DNA (cDNA) by the enzyme reverse transcriptase, which is required because DNA polymerase cannot use RNA as template.
After cDNA is synthesised, The double-stranded cDNA is denatured into single-stranded templates, so that primers can anneal. The temperature is raised to separate strands and lowered for annealing.
In annealing phase, primers bind to complementary sequences on the ssDNA-templates, which is governed by nucleic acid hybridization principle; specificity is determined by primer design.
DNA polymerase is used to elongate from the 3’ end of each primer, nucleotides are added sequentially, so that new DNA strand (complementary to template) is produced by replication-like action.
The cycles of denaturation, annealing, and extension (elongation) are repeated many times (eg ~25-40 cycles), so exponential amplification of target DNA region is achieved.
During the reverse transcription step, either random primers, oligo(dT) primers, or gene-specific primers are used to initiate synthesis of cDNA, depending on type of RNA target and desired specificity.
In variant forms, The entire process (reverse transcription + PCR) is performed in single tube (“one-step RT-PCR”) or in separate steps (“two-step RT-PCR”), which affects handling, sensitivity, flexibility.910
Requirements of Reverse Transcription PCR (RT-PCR)
- RNA Sample must be provided, because RT-PCR uses RNA as starting template, not DNA; mostly messenger RNA (mRNA) is used, and sample RNA is converted into cDNA before any amplification. (from reference + confirmed elsewhere)
- Reverse Transcriptase Enzyme is required – an enzyme that catalyses formation of complementary DNA (cDNA) from the RNA template. Its properties such as thermostability, RNase H activity, fidelity, processivity are important because they affect yield & quality of cDNA.
- DNA Polymerase Enzyme must be included, for amplification of cDNA. Thermostable polymerases (eg Taq DNA Polymerase) are often used so that repeated heating/cooling cycles do not denature it, enabling many PCR cycles.
- Primers are needed:
- Random Primers (short oligonucleotides, 6-9 bases) are used to bind at multiple sites along RNA, useful when sample RNA has secondary structure or when full coverage is desired.
- Oligo(dT) Primers (≈12-18 nucleotides, strings of deoxythymidine) are used to anneal to poly(A) tail of mRNA, ensuring priming at transcript’s 3′ end.
- Sequence-specific Primers are used when a specific region or gene is to be amplified, often in one-step RT-PCR, for higher specificity.
- Deoxynucleotide Triphosphates (dNTPs) must be present, as the building blocks (dATP, dGTP, dTTP, dCTP) for synthesis of cDNA and new DNA strands during amplification. Concentrations and purity affect efficiency and error rates.
- Buffer & Other Chemicals are necessary:
- Reaction buffer with correct salt concentration, pH, stabilisers must be used to maintain enzyme activity and fidelity.
- Cofactors, especially Mg^2+ (sometimes other divalent ions), must be included since they are required by both reverse transcriptase & DNA polymerase for catalysis.
- Thermocycler (PCR Machine) is required so that thermal cycling steps (denaturation, annealing, extension) can be regulated precisely and repeated for many cycles, leading to exponential amplification of target DNA.
Types of Reverse Transcription PCR (RT-PCR)
- One-Step RT-PCR –
- The reverse transcription and PCR amplification are combined in a single tube, under one buffer system, which simplifies handling and reduces risk of contamination.
- Gene-specific primers only are used in the reverse transcription step, because multiplex or other priming strategies are limited by combined reaction conditions.
- High throughput applications are supported, when many samples are needed, because time and pipetting steps are reduced.
- Sensitivity or flexibility is reduced in some cases, because optimization of RT and PCR steps cannot be done separately.
- Two-Step RT-PCR –
- The reverse transcription is done first, then the PCR amplification of the resulting cDNA is performed in a separate tube, so that buffers / enzymes for each step can be optimized independently.
- Primers of different kinds (oligo(dT), random hexamers, gene-specific) are permitted for RT step, because separate RT reaction allows choice.
- Multiple targets can be amplified from the same cDNA pool, which increases efficiency when many genes are to be studied.
- Risk of contamination is increased, because additional handling (tube transfer etc.) is involved.
- Real-Time RT-PCR (RT-qPCR) –
- Amplification is monitored during the PCR cycles, by fluorescent signals, so that quantification is done in real time, not only at end-point.
- SYBR Green dyes or probe-based systems (TaqMan etc.) are used for fluorescence detection, which allow for both specificity and quantification.
- End-Point RT-PCR –
- The PCR products are analysed after the cycles are completed, by gel electrophoresis or other detection methods, which gives qualitative or semi-quantitative results.
- Comparative, competitive, and relative quantification methods are included under end-point RT-PCR, when quantification by comparison is done after amplification.
- Relative Quantification RT-PCR –
- Expression of target gene is compared to that of internal control (housekeeping gene), so fold-change is assessed rather than absolute copy number.
- Multiple samples are compared, normalization is done using control gene, variation is reduced across samples.
- Absolute / Comparative / Competitive Quantification RT-PCR –
- An external standard curve or a known competitor RNA is included, so that absolute copy number of target RNA may be determined.
- In competitive RT-PCR, competitor is designed to amplify under same conditions but distinguishable (by size or sequence), which allows quantification.
- Isothermal RT Amplification methods (related / variant) –
- RT-LAMP (Reverse Transcription Loop-mediated Isothermal Amplification) is used, where amplification occurs at constant temperature (≈ 60-65°C), instead of thermocycling, which isothermal method is more rapid in some settings
- Such variant methods are sometimes used for pathogen detection when simplicity and speed are required, particularly in field or low resource settings.1112
What is One-Step RT-PCR?
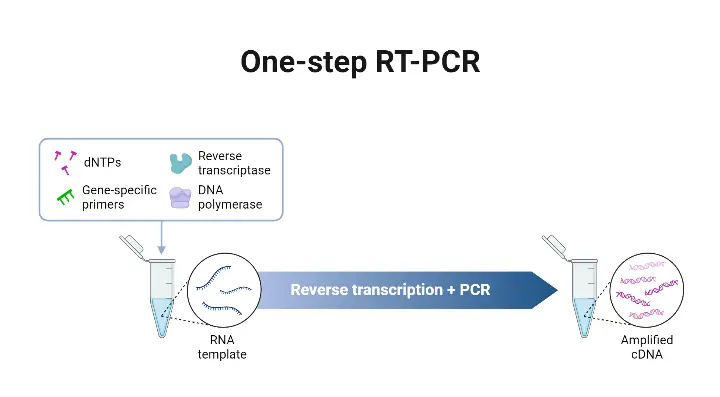
Definition –
- A reaction format is used in which reverse transcriptase (RT) and DNA polymerase are combined in same tube, same buffer, so that cDNA synthesis and PCR amplification are done consecutively without opening tube.
- Gene-specific primers are used, because universal primers or random primers are not feasible for RT in one-step set-up, as conditions are fixed for both RT and PCR.
Properties / Inherent Features –
- High throughput suitability is present, since multiple samples can be processed rapidly, with less handling.
- Minimal sample manipulation is required, which reduces risk of contamination and pipetting error.
Advantages –
- Workflow is simplified, because two enzymatic reactions (RT + PCR) are merged, so fewer pipetting steps are needed.
- Time consumed / bench time is reduced, which allows faster turnaround.
- Variation between reactions is lowered, because same tube, same environment, reaction conditions held constant.
Limitations / Disadvantages –
- Sensitivity is sometimes lower, since reaction conditions must compromise between optimal reverse transcription and optimal PCR; so neither step is ideal.
- Flexibility is reduced, because only gene-specific primers are usable, and no cDNA stock is created for later reuse of other targets.
- Troubleshooting is more difficult, because if something fails, it is unclear whether the RT or PCR component is at fault; separate optimisation is not possible.
- More starting RNA may be required, when multiple targets are to be tested or when template quality is poor.
Applications / Suitable Use Cases –
- High-throughput screening where only a few known genes are to be examined in many samples is ideal.
- Diagnostic assays where speed and reduced handling / contamination risk are important are suited.
- Situations where consistent, reproducible amplification of same target is required and experimental conditions / primers are well validated.1314
What is Two-Step RT-PCR?
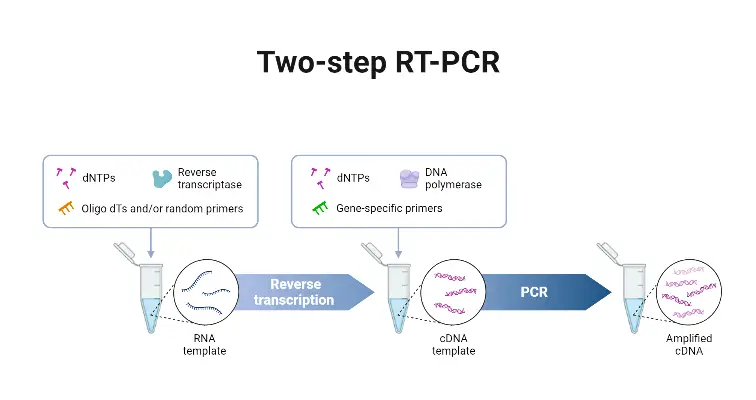
Definition –
- Two-Step RT-PCR is a format in which reverse transcription (RT) and PCR amplification are done in separate reactions, in different tubes, so that cDNA is first synthesized, then amplified.
- The priming strategy for the RT part is flexible; oligo(dT), random hexamers, or gene-specific primers may be used
Properties / Inherent Features –
- A stable cDNA pool is made, which may be stored for further PCRs’ use, from same RNA sample.
- Optimization of buffer / enzymes / reaction conditions for RT step and PCR step can be done independently, which improves efficiency / sensitivity.
Advantages –
- Multiple target genes can be amplified from same cDNA, so many assays may be run, from one RT reaction.
- Higher sensitivity and better yield may be achieved, because each reaction (RT, PCR) can be tuned for its own optimum, when RT and PCR are separated.
- Flexibility of primer usage in RT (random hexamers / oligo(dT)) allows broader representation of transcripts, not only those with known sequences.
- Comparisons across experiments are made easier, since same cDNA pool may be used for internal controls and target genes, reducing variation due to RT step.
Limitations / Disadvantages –
- More hands-on time is demanded, because two separate reactions must be set up / handled
- Contamination risk is increased, due to more pipetting / open tubes, cDNA transfer between tubes.
- Workflow is slower, which may not be ideal for high throughput settings.
- Reagents cost may be higher / resource use more, because separate reaction mixes etc are needed
Applications / Situations when Two-Step RT-PCR is Preferred –
- When RNA sample is limited, and greater sensitivity / detection of low abundance transcripts is required.
- When the number of genes to be tested is many, including those not known at the beginning, so that making cDNA archive is useful.
- When challenging sequences (e.g. high GC content, secondary structure) are involved, that require fine optimization of RT / PCR conditions separately.1516
Differences between Two-Step RT-PCR and One-Step RT-PCR
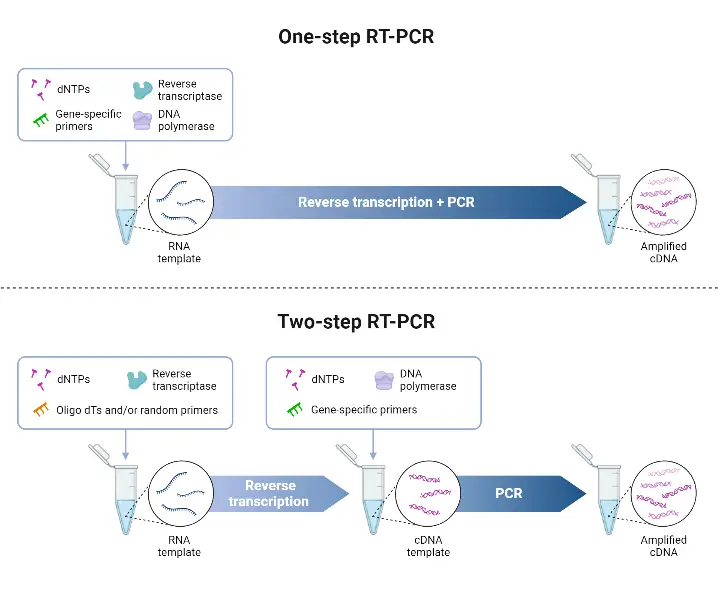
| Feature | One-Step RT-PCR | Two-Step RT-PCR |
|---|---|---|
| Setup / Workflow | Both reverse transcription (RT) and PCR are performed in same tube / single reaction mix, under conditions compatible for both steps. | RT and PCR are done in separate tubes / separate reactions, each optimized independently. |
| Primers used for RT | Only sequence-specific primers are used. | Flexible priming: oligo(dT), random hexamers, or gene-specific primers may be used |
| Number of targets / genes per sample | Fewer targets are practical, because only specific primers are used, and multiplexing etc is more difficult. | Many targets per sample can be tested, because cDNA generated can be used for multiple PCRs. |
| Throughput / speed | Faster, fewer handling steps, good for high throughput | Slower, more pipetting, more sample handling; more time needed. |
| Sensitivity / Efficiency | Sometimes lower sensitivity, because reaction conditions are compromise between RT & PCR, optimization of each step is constrained | Higher sensitivity can be achieved, because RT and PCR steps can be individually optimised |
| Risk of contamination / variation | Lower risk of sample contamination / variation, due to fewer transfers and handling. | Higher risk, because of multiple steps, open tubes, transfers etc |
| cDNA storage / reuse | cDNA not separately generated for storage / reuse; entire reaction is used for PCR. | cDNA pool is made, can be stored, reused for multiple PCR targets |
| Flexibility in reaction optimization | Lower flexibility, since RT & PCR must share buffer / conditions, and reaction must be compromise. | Higher flexibility: buffer, enzyme, temperature etc can be optimized separately for RT and for PCR. |
Steps/Procedure of Reverse Transcription PCR (RT-PCR)
- Preparatory Stage –
- Sample RNA is extracted / purified from biological material, using methods ensuring integrity, removal of inhibitors
- Genomic DNA is removed (by DNase treatment or design of primers across exon-exon junctions), which prevents non-specific amplification.
- Primers are designed / synthesized. Primer design is done such that specificity, melting temperature, absence of self-complementarity, minimal primer dimer formation are considered.
- Reaction components (reverse transcriptase, DNA polymerase, dNTPs, buffer, Mg2+ etc.), and the thermal cycler are prepared / equilibrated.
- Reverse Transcription Step –
- Primer (gene-specific / random hexamer / oligo(dT) depending on type) is annealed to RNA, when temperature permitted, so that reverse transcriptase can bind.
- Reverse transcriptase enzyme is used to synthesize complementary DNA (cDNA) from RNA template, under suitable temperature (often ~40-50°C, for a duration e.g. 10-30 minutes) depending on enzyme.
- PCR Amplification Step –
- Thermal cycling is applied: Denaturation of cDNA, annealing of primers to cDNA, extension by DNA polymerase. These cycles are repeated many times (often 25-40 cycles) to amplify the target DNA.
- Annealing temperature is optimized based on primer melting temperature. Extension temperature is set for DNA polymerase optimum. Denaturation temperature (usually ~94-95°C) is used.
- In one-step RT-PCR, amplification follows immediately in same reaction; in two-step RT-PCR, cDNA is used in separate amplification reaction.
- Product Analysis Stage –
- Amplified products are analysed, by gel electrophoresis, if end-point detection is used. Bands are visualized (e.g. ethidium bromide, or other stains) to confirm size and presence.
- If real-time RT-PCR is used, amplification is monitored during the PCR cycles via fluorescence, so product analysis is done in real time rather than at end-point.
- Data (threshold cycles, melt curves etc) are interpreted, replicates are compared, internal controls are used to validate that reaction worked.1718
Applications of Reverse Transcription PCR (RT-PCR)
- Gene Expression Analysis –
- The expression level of target genes is measured, when mRNA abundance is needed, even if transcripts are in low amount.
- Differential expression under different conditions (stress, treatment, developmental stage etc.) is compared.
- Transcript Variant / Splice Variant Detection –
- Alternative splicing forms are detected, by designing primers across exon-junctions, which allow discovery of transcript variants.
- Isoforms differing by small insertions / deletions / inclusion/exclusion of exons are discriminated
- Pathogen Detection / Diagnostics –
- Viral RNA (e.g. viruses causing infectious diseases) is detected, so diagnosis is enabled even before antibodies are produced.
- Detection of genetically modified organisms (GMOs) via specific RNA signatures is performed.
- Validation of RNA Interference (RNAi) or Gene Knockdown –
- The effect of gene silencing (by RNAi) is quantified, by comparing levels of mRNA before and after treatment.
- cDNA Template Generation for Cloning / Sequencing –
- The cDNA produced by RT is used for downstream experiments (cloning, sequencing) when the RNA template is converted.
- Absolute / Relative Quantification of RNA –
- The number of copies / fold changes of RNA transcripts are quantified, using RT-qPCR (real-time).
- Monitoring Pathogen Load / Infectious Disease Progression –
- The load of virus or bacteria in a patient is monitored over time by quantifying pathogen RNA, which is useful for treatment decisions.
- Basic / Biomedical Research –
- Studies in molecular biology, developmental biology, cell signalling etc are conducted, where measurement of gene transcripts (mRNA) is needed
- Expression profiling under perturbation (drug, mutation) is performed.19202122
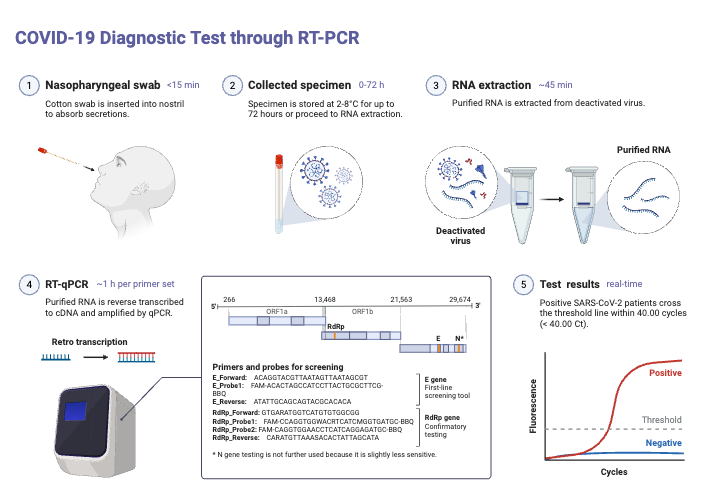
Advantages of Reverse Transcription PCR (RT-PCR)
- The Sensitivity is Very High, enabling detection of very small amounts of RNA, in some cases from single cells.
- A Large Dynamic Range is Provided, so that low-abundance and high-abundance transcripts may be quantitated in same experiment.
- The Specificity is Ensured, due to use of sequence-specific primers (and probes) which reduce non-specific amplification.
- Turnaround Time is Reduced, because real-time RT-PCR allows monitoring during cycles rather than after end-point, eliminating post-PCR processing like gel electrophoresis.
- Small Sample Amounts are Needed, when only little RNA is available, RT‐PCR can be done reliably.
- Multiplexing Capability is Enabled, several targets can be amplified in same reaction tube (if properly designed) to save time / reagents.
- Wide Applicability is Furnished, for diagnostics, gene expression studies, pathogen detection etc.
- Reproducibility is Achieved, because once optimized, results can be similar across runs / labs, when proper controls are included.23242526
Limitations of Reverse Transcription PCR (RT-PCR)
- The Quality of RNA must be very high, because degraded RNA leads to false negatives or poor results, which can mislead an analysis.
- The Requirement for prior knowledge of target sequence is imposed, since primers / probes must be designed for specific regions, which limits detection of new or unexpected variants.
- Specialized Equipment and Highly-Trained Personnel are needed, which makes cost high and accessibility limited, especially in resource-poor settings.
- Inhibition of Amplification may happen, because contaminants / inhibitors in sample (e.g. from extraction) interfere, which causes false negatives or under-quantification.
- Standardization is Difficult, because different labs use different protocols, reagents, housekeeping genes etc., so reproducibility / comparability is impaired.
- The Dynamic Range / Quantification at end-point is Unreliable, because when exponential amplification proceeds into plateau phase, linearity is lost, which causes inaccurate quantification.
- Multiplexing is Limited, because cross-reactivity / overlapping spectra of dyes or probes, and complexity of designing multiple primer sets correctly hinder multiplex assays.
- False Negatives or Variable Sensitivity are Produced, because sample collection, handling, RNA quality, viral load etc vary, which causes test to miss some cases.
- Cost / Scalability Problems are Presented, because reagents, instruments, and containment / infrastructure are expensive, which restricts scaling in many settings.
- Unknown or Highly Variable Sequences may be Missed, because primers designed for known sequences fail when mutation / variation occurs, which reduces detection of new strains.23242526