What is a Retrovirus?
- Retroviruses are unique, enveloped, positive-stranded RNA viruses. They exhibit a spherical shape and have a distinct mode of replication. The key feature of retroviruses is the presence of an enzyme called reverse transcriptase. This RNA-dependent DNA polymerase enables the virus to create DNA from its RNA genome, hence the name retrovirus, meaning “reverse.”
- Upon entering a host cell, the retrovirus uses reverse transcriptase to generate a DNA copy of its RNA genome. Initially, this results in an RNA-DNA hybrid, which then becomes double-stranded DNA. This viral DNA, known as a provirus, integrates into the host cell’s DNA, becoming a permanent part of the host’s genetic material.
- Unlike typical genetic transcription, which goes from DNA to RNA to proteins, retroviruses reverse this process by converting RNA to DNA. This integration into the host’s genome allows the provirus to replicate along with the host cell, ensuring the viral genes are perpetuated.
- The discovery of reverse transcriptase was groundbreaking, first demonstrated by scientists Baltimore and Temin. Their work showed that retroviruses replicate through a DNA intermediate, a process previously unknown.
- Retroviruses encompass several subfamilies. Oncoretroviruses, like the human T-lymphotropic virus (HTLV), can cause cancers such as leukemia. Lentiviruses, including HIV-1 and HIV-2, are responsible for AIDS. Another group, spumaviruses, are benign and not associated with diseases in humans or animals.
- The specialized enzymes of retroviruses have made them valuable tools in molecular biology. They are extensively used in gene delivery systems for research and therapeutic purposes. Endogenous retroviruses, remnants of ancient infections, provide evidence that retroviruses have been infecting vertebrates for at least 450 million years.
Definition of Retrovirus
A retrovirus is a type of RNA virus that replicates by converting its RNA genome into DNA through the enzyme reverse transcriptase. This DNA is then integrated into the host cell’s genome, where it directs the production of new viral particles. Retroviruses are associated with various diseases, including certain cancers and HIV/AIDS.
Classification of Retrovirus
Retroviruses are categorized based on transmission modes, host range, morphology, and biological properties. This classification is crucial for understanding their behavior, pathogenicity, and application in research.
1. Classification by Transmission Mode
- Exogenous Retroviruses
- Definition: These viruses are transmitted between individuals. They are infectious and can spread through horizontal transmission.
- Baltimore Classification:
- Group VI: Single-stranded RNA viruses with a DNA intermediate in their life cycle.
- Group VII: Double-stranded DNA viruses with an RNA intermediate in their life cycle.
- Endogenous Retroviruses
- Definition: These viruses are inherited through germ cells and are integrated into the chromosomes of sperm or oocytes. They are not typically infectious in their host species but can infect other species.
- Classification:
- Class I: Similar to gammaretroviruses.
- Class II: Similar to betaretroviruses and alpharetroviruses.
- Class III: Similar to spumaviruses.
2. Classification by Host Range
- Xenotropic Retroviruses
- Definition: These viruses replicate only in cells of a different species from their origin.
- Examples: Generally endogenous retroviruses.
- Ecotropic Retroviruses
- Definition: These viruses replicate only in cells of the host species and closely related species.
- Examples: Certain exogenous retroviruses.
- Amphotropic Retroviruses
- Definition: These viruses can replicate in both the host species and heterologous cells.
- Examples: Some exogenous retroviruses.
3. Classification by Morphology
- Type A
- Characteristics: Found intracellularly and not infectious.
- Examples: Rarely encountered in laboratory settings.
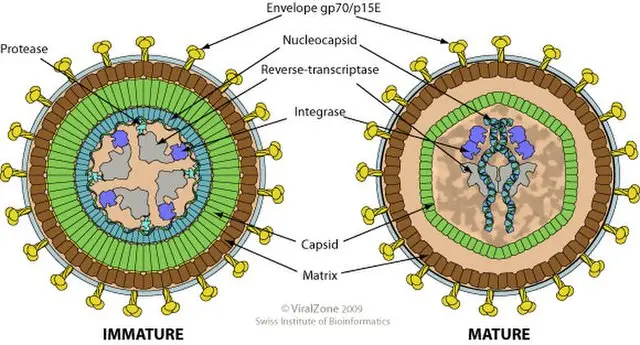
- Type B
- Characteristics: Slightly larger than type C, with an eccentrically located core. Assembles its core in the cytoplasm and acquires its envelope as it buds from the plasma membrane.
- Examples: Mouse mammary tumor virus (MMTV).
- Type C
- Characteristics: Most common, with particles forming by budding from the plasma membrane. These are 80-120 nm in diameter with a centrally located core.
- Examples: Many known retroviruses.
- Type D
- Characteristics: Similar in assembly to type B but closely resembles type C in morphology.
- Examples: Some viruses with less distinct features.
4. Classification by Biological Properties
- Oncovirinae
- Characteristics: Oncoviruses are associated with cancer and malignant tumors.
- Examples: Human T-lymphotropic viruses (HTLV).
- Lentivirinae
- Characteristics: Lentiviruses are associated with slowly progressing inflammatory and degenerative diseases.
- Examples: Human immunodeficiency viruses (HIV-1, HIV-2).
- Spumavirinae
- Characteristics: Spumaviruses cause foamy degeneration of cells in vitro but have not been linked to diseases in animals or humans.
- Examples: Various foamy viruses.
Structure of Retrovirus
The structure of retroviruses is characterized by several distinct components, each playing a crucial role in their life cycle and function. Here is a detailed examination of their structure:
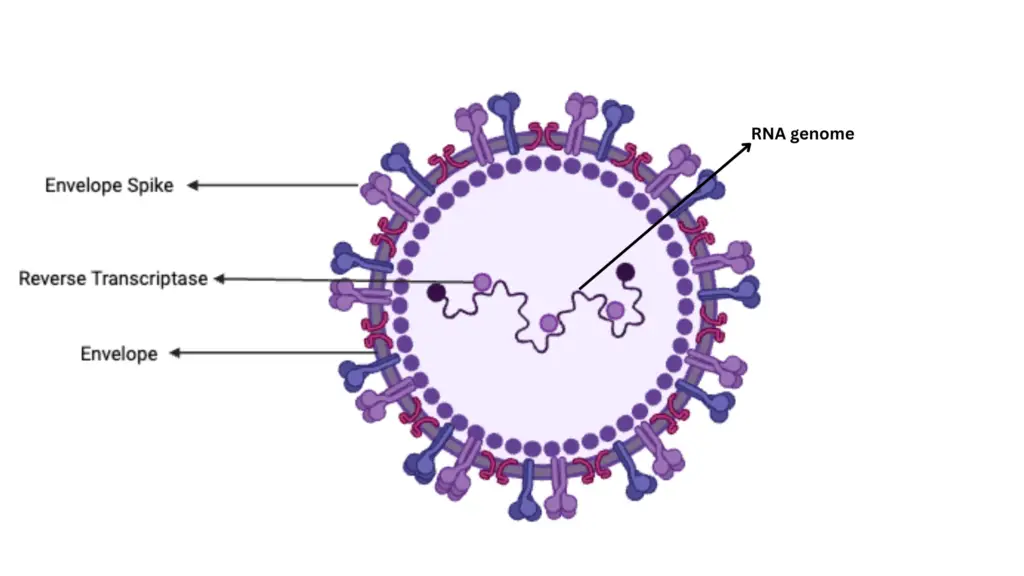
1. General Structure
- Size: Retroviruses measure approximately 80-130 nm in diameter.
- Components:
- Core (Nucleoid): Contains ribonucleoprotein and is surrounded by a protein capsid.
- Envelope: A lipid membrane derived from the host cell’s plasma membrane, embedded with viral glycoproteins.
2. Envelope
- Composition:
- Lipids: Derived from the host cell’s membrane.
- Glycoproteins: Encoded by the viral env gene.
- Functions:
- Protection: The lipid bilayer shields the virus from the external environment.
- Cell Entry/Exit: Facilitates the virus’s ability to enter or exit host cells through endosomal membrane trafficking or direct fusion with the host cell membrane.
3. Core (Nucleoid)
- Composition:
- Capsid: A protein shell encasing the viral RNA.
- Inner Coat Protein: Positioned between the capsid and the envelope.
- Components:
- RNA: Two identical single-stranded RNA molecules, each 7–10 kilobases long, form a dimer through base pairing.
- Proteins:
- Gag Proteins: Major components of the viral capsid, including matrix (MA) and nucleocapsid (NC) proteins, which are crucial for packaging the RNA.
- Protease (PR): Cleaves precursor proteins during virion maturation.
- Pol Proteins: Involved in the synthesis of viral DNA and its integration into the host genome.
- Env Proteins: Comprise the surface (SU) and transmembrane (TM) proteins, responsible for virus binding and entry into host cells.
4. RNA Structure
- Genomic RNA (gRNA):
- Cap and Poly(A) Tail: The RNA has a 5′ cap and a 3′ poly(A) tail, crucial for stability and translation.
- Terminal Noncoding Regions:
- 5′ End: Includes R (repeated sequence), U5 (unique sequence), PBS (primer binding site), and L (leader region).
- 3′ End: Includes PPT (polypurine tract), U3 (unique sequence), and R (repeated sequence).
- Functions: Essential for reverse transcription, packaging, and replication.
5. Enzymes
- Reverse Transcriptase (RT): An RNA-dependent DNA polymerase that synthesizes DNA from the RNA genome.
- RNase H: Cleaves RNA during reverse transcription, essential for converting RNA to DNA.
- Integrase: Facilitates the integration of viral DNA into the host genome.
- Protease: Assists in the maturation of viral proteins.
6. Additional Features
- Buoyant Density: Retroviruses have a density of 1.16-1.18 g/ml in sucrose.
- Sensitivity: Sensitive to lipid solvents and detergents, but resistant to ultraviolet light and gamma irradiation.
- Composition: Typically contain 60%-70% protein, 30%-40% lipid, 2%-4% carbohydrate, and 1% RNA.
Genomic Structure of Retrovirus
Retroviruses exhibit a unique genomic structure that is crucial for their replication and pathogenicity. Here is an in-depth exploration of their genomic architecture:
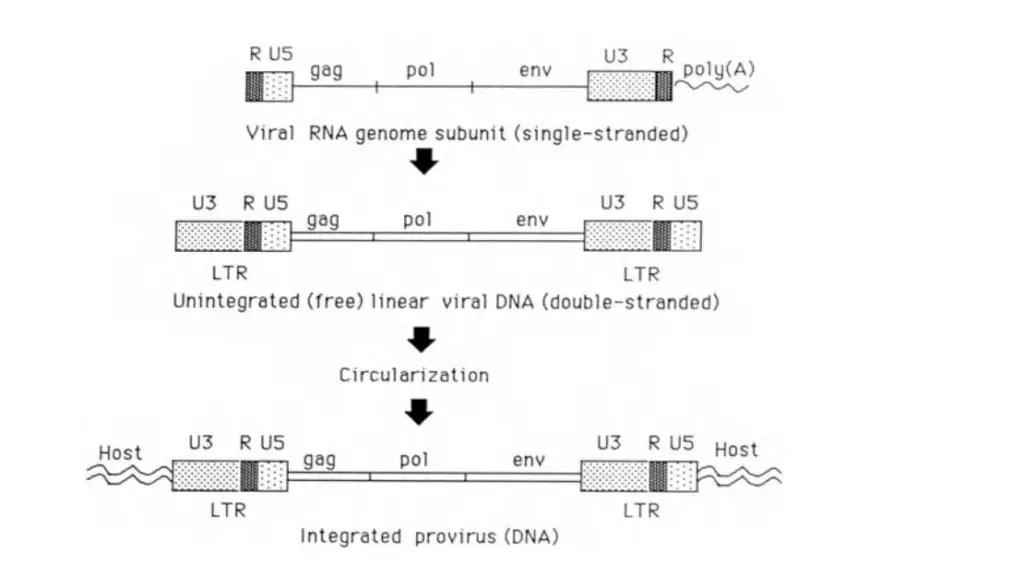
1. Genome Composition
- RNA Type: Retroviral genomes are composed of positive-sense, single-stranded RNA. This RNA is similar to the mRNA found in eukaryotic cells.
- Structure: The RNA molecules are capped at the 5′ end and polyadenylated at the 3′ end, mimicking the structure of cellular mRNA. Each viral particle contains two identical RNA molecules linked at their 5′ ends by noncovalent bonds, giving the genome a diploid nature.
2. Genomic Layout
- Gene Order: The typical retroviral genome is organized in a 5’–gag–pol–env–3′ arrangement.
- Gag: Encodes core proteins that form the structural components of the viral capsid.
- Pol: Encodes reverse transcriptase, which is essential for converting viral RNA into DNA, as well as other enzymes like protease and integrase.
- Env: Encodes the envelope proteins responsible for virus entry into host cells.
- Regulatory Sequences: Flanking the coding regions are regulatory sequences:
- 5′ End: Contains R (repeat) and U5 (unique sequence 5′ of the gag gene).
- 3′ End: Contains U3 (unique sequence 3′ of the env gene) and R (repeat).
3. Long Terminal Repeats (LTRs)
- Formation: During reverse transcription, the R and U5 sequences at the 5′ end and U3 and R sequences at the 3′ end are duplicated. This duplication forms identical LTRs at both ends of the viral DNA.
- Function: LTRs play a crucial role in integrating the viral DNA into the host genome and regulating transcription.
4. Replication Competence and Transformation
- Replication-Competent Viruses:
- Structure: These viruses possess all three essential genes (gag, pol, and env) and are capable of replication.
- Transformation: They do not cause malignant transformation of cultured cells. Hence, they are termed replication-competent but transformation-defective.
- Transformation-Competent Viruses:
- Genetic Modification: Some retroviruses acquire genes of cellular origin (cellular oncogenes, c-onc) through recombination. These genes integrate into the viral genome, resulting in transformation-competent viruses.
- Impact: These viruses are capable of inducing malignant transformation in cultured cells but are replication-defective if the oncogene is integrated into a replicative gene.
- Rescue Mechanism: They can replicate if co-infected with a replication-competent virus, which provides the necessary proteins for encapsidation of the defective genome. This process allows the production of replication-competent viruses from cultures containing both types.
5. Isolation and Study
- V-One Positive Retroviruses: These viruses can only be isolated from cultures that also produce replication-competent viruses. This is due to their inability to replicate independently without the assistance of helper viruses.
What are the Retroviral Proteins?
Retroviral proteins are essential for the virus’s structure, replication, and infectivity. Each class of proteins plays a specific role, and their functions are crucial for the virus’s lifecycle. Below is a detailed overview of the main retroviral proteins:
- Gag Proteins
- Matrix (MA):
- Function: MA is a structural protein found on the inside of the virus envelope. It is derived from the amino-terminal domain of the Gag polyprotein.
- Modification: Most MA proteins are myristylated at the Gly residue in position 2. In avian sarcoma/leukemia virus (ASLV), MA is also acetylated at the initiating methionine.
- Capsid (CA):
- Function: CA is the principal structural protein of the virion core, derived from the central region of the Gag polyprotein.
- Role: CA forms the core structure that encases the viral RNA and essential proteins.
- Nucleocapsid (NC):
- Function: NC is a small, basic protein derived from the carboxy-terminal domain of the Gag polyprotein. It tightly binds to the viral genomic RNA.
- Role: NC forms a ribonucleoprotein complex within the core, playing a critical role in RNA encapsidation.
- Matrix (MA):
- Pro Proteins
- Protease (PR):
- Function: PR is an aspartyl-protease that cleaves the Gag, Gag-Pol, and Gag-Pol-Env polyproteins into their mature forms. It is also involved in removing carboxy-terminal fragments of Env in some viruses, such as MLV.
- Structure: Retroviral proteases function as homodimers, essential for the maturation of viral proteins.
- dUTPase (DU) in Type-B and Type-D Retroviruses:
- Function: DU is not essential for viral replication in vitro but helps manage cellular dUTP levels.
- Relation: The DU encoded by pro is distinct from the dUTPase found in some lentiviruses.
- Protease (PR):
- Pol Proteins
- Reverse Transcriptase (RT):
- Function: RT is a DNA polymerase that can copy RNA or DNA templates and possesses RNase H activity. It initiates minus-strand DNA synthesis using a cellular tRNA primer and plus-strand DNA synthesis using a primer that resists RNase H.
- Structure: RT in avian type-C and HIV-1 viruses exists as heterodimers, whereas some retroviruses have monomeric RT.
- Integrase (IN):
- Function: IN is responsible for processing the long terminal repeats (LTRs) of the retroviral genome and integrating the linear double-stranded DNA into the host cell DNA, forming a provirus.
- Role: This enzyme is crucial for establishing a persistent viral infection.
- dUTPase (DU) in Certain Lentiviruses:
- Function: Similar to the DU in Type-B and Type-D viruses, this enzyme helps manage dUTP levels but is not essential for replication in vitro.
- Reverse Transcriptase (RT):
- Env Proteins
- Surface (SU):
- Function: SU mediates viral adsorption by binding to specific cell surface receptors. It is encoded in the amino-terminal region of the env gene and is translated from spliced subgenomic mRNA.
- Modification: SU proteins are modified by N-linked glycosylation and are separated from the Transmembrane (TM) protein by cellular cleavage.
- Transmembrane Envelope (TM):
- Function: TM facilitates virus entry by triggering fusion between the virus and host cell membranes. It is encoded in the carboxy-terminal region of the env gene.
- Modification: TM proteins are also modified by N-linked glycosylation, aiding in their role in virus entry and membrane fusion.
- Surface (SU):
- Accessory Proteins
- Lentivirus Proteins:
- Tat: Activates transcription by binding to TAR, enhancing viral gene expression.
- Rev: Facilitates the transport of unspliced and incompletely spliced mRNAs to the cytoplasm.
- Nef: Reduces CD4 levels on the cell surface and stimulates infected cells to divide.
- Vpr: Causes cell cycle arrest in G2 and aids in the transport of the preintegration complex into the nucleus.
- Vpu: Degrades newly synthesized CD4 and assists in viral assembly and release.
- Vif: Aids in producing infectious virions by enhancing their stability.
- HTLV/BLV Proteins:
- Tax: Stimulates transcription by binding to specific response elements in the LTR.
- Rex: Facilitates the transport of unspliced RNAs to the cytoplasm.
- Spumavirus Proteins:
- Tas: A trans-activator that stimulates transcription by binding to the U3 region of the LTR.
- Bet: A protein of unknown function, expressed at high levels but not essential for replication.
- B-type Virus Proteins:
- Sag: Interacts with T-cell receptors to induce activation signals, affecting T-cell subsets.
- Fish Virus Proteins:
- ORF-a and ORF-b: Encode proteins related to cellular cyclins, impacting viral replication.
- Lentivirus Proteins:
Regions and Features of Replication-competent Retroviral RNA
Replication-competent retroviral RNA contains specific regions and features essential for viral replication and function. Below is an overview of these regions, each playing a crucial role in the virus’s lifecycle.
- Cap Site
- Function: The cap site is located at the 5′ end of the viral RNA subunit. It is linked to a methylated “cap” ribonucleotide by a 5′-5′ linkage.
- Role: This cap structure is vital for RNA stability and translation initiation.
- Repeat (R)
- Function: The R region is a short sequence, typically 15–250 nucleotides, repeated at both ends of the genomic RNA. It marks the boundaries for RNA transcription initiation and polyadenylation.
- Features:
- Polyadenylation Signal: In many retroviruses, R includes the polyadenylation signal sequence (AAUAAA), which is crucial for adding the poly(A) tail.
- TAR (Tat-responsive)
- Function: In primate lentiviruses, TAR is a highly structured region within R that serves as the binding site for the Tat trans-activator.
- Role: TAR is essential for regulating viral gene expression by enhancing transcription.
- U5 (Unique 5′)
- Function: U5 is a sequence of 70–250 nucleotides located between R and the primer-binding site (PBS).
- Features:
- Presence: U5 appears once in genomic RNA and twice in viral DNA as part of the long terminal repeat (LTR).
- PBS (Primer-binding Site)
- Function: The PBS is usually 18 nucleotides long, beginning with 5′ TGG. It is adjacent to U5 and complementary to the 3′ terminus of a specific host tRNA species.
- Role: PBS is the binding site for a tRNA that acts as a primer for reverse transcriptase, initiating the synthesis of the minus (–) strand of viral DNA.
- gag
- Function: This coding domain is the first of four in the genomes of replication-competent retroviruses. It encodes the Gag polyprotein, which is cleaved into major structural proteins: matrix (MA), capsid (CA), and nucleocapsid (NC).
- Exceptions: In spumaviruses, Gag proteins are not extensively cleaved.
- pro
- Function: The second coding domain encodes part of the Gag-Pro or Gag-Pro-Pol polyprotein. The cleavage products include protease (PR) and, in some viruses, dUTPase (DU).
- Role: PR is crucial for the maturation of viral proteins.
- pol
- Function: The third coding domain encodes part of the Gag-Pro-Pol polyprotein. Cleavage produces reverse transcriptase (RT) and integrase (IN), and, in some lentiviruses, dUTPase (DU).
- Expression: In spumaviruses, pol is expressed via a spliced mRNA as a Pol-Pol polyprotein.
- env
- Function: The final coding domain encodes the Env polyprotein, which is cleaved into surface (SU) and transmembrane (TM) proteins. These are integral to the viral envelope.
- Role: SU and TM are essential for viral entry and envelope structure.
- SD (Splice Donor Site)
- Function: This site allows the joining of an upstream (5′) portion of viral RNA to a downstream (3′) portion at the splice acceptor (SA), forming spliced, subgenomic mRNA.
- Role: It facilitates the production of different mRNA species necessary for various viral proteins.
- SA (Splice Acceptor Site)
- Function: The SA site enables the joining of a downstream (3′) portion of viral RNA to an upstream (5′) portion at the splice donor (SD).
- Role: It is crucial for generating spliced mRNA.
- RRE (Rev or Rex Response Element)
- Function: RRE is a highly structured sequence in lentivirus (or HTLV-BLV) RNAs. It serves as a binding site for the Rev or Rex protein.
- Role: RRE aids in the transport of unspliced or partially spliced RNAs from the nucleus to the cytoplasm.
- CTE (Constitutive Transport Element)
- Function: CTE is found in D-type and ASLV RNAs and promotes the transport of unspliced RNAs from the nucleus to the cytoplasm.
- Role: It operates in the absence of virus-coded proteins such as Rev or Rex, facilitating RNA export.
- PPT (Polypurine Tract)
- Function: The PPT is a purine-rich sequence of 7–18 nucleotides located upstream of U3.
- Role: It is cleaved during reverse transcription to produce the RNA primer for synthesis of the plus (+) strand of viral DNA. Some retroviruses use multiple PPTs to prime DNA synthesis at various sites.
- U3 (Unique 3′)
- Function: U3 is a sequence of several hundred nucleotides positioned between PPT and R near the 3′ end of viral RNA.
- Features:
- Presence: U3 is found once in viral genome RNA and twice in viral DNA as part of the LTR.
- Role: U3 contains promoter-enhancer sequences that regulate viral RNA transcription.
- Poly(A) Tract
- Function: The poly(A) tract is a homopolymer of 50–200 adenylic acid residues located at the 3′ end of the viral RNA following the R sequence.
- Role: This tract is added posttranscriptionally and is crucial for RNA stability and export. A signal for polyadenylation (AAUAAA) is generally present upstream of the poly(A) tract.
Regions and Features of Retroviral DNA
Retroviral DNA exhibits distinct regions and features essential for its replication and integration into the host genome. Below is a detailed explanation of these components:
- LTR (Long Terminal Repeat)
- Description: LTRs are regions composed of several hundred base pairs (∼300–1800) and consist of three main segments: U3, R, and U5, arranged from 5′ to 3′.
- Location and Structure:
- Unintegrated and Integrated DNA: LTRs are present at both ends of the unintegrated and integrated (proviral) linear DNA. They can also be found in closed circular forms of retroviral DNA, which may contain one or two LTRs.
- Proviral DNA Modifications: In proviral DNA, the 5′ LTR is missing two nucleotides from the 5′ end of U3, and the 3′ LTR is missing two base pairs from the 3′ end of U5.
- Function: LTRs play a crucial role in the regulation of viral gene expression and integration. They contain promoter and enhancer elements essential for transcription.
- IR (Inverted Repeat; Also Called the att Site)
- Description: IRs are short sequences, typically ∼3–25 base pairs, that form perfect or slightly imperfect inverted repeats at the ends of the LTR.
- Function:
- Recognition Sites: These inverted repeats provide recognition sites for integrase, the enzyme responsible for integrating the viral DNA into the host genome.
- Proviral DNA Modifications: In proviral DNA, two base pairs are missing from the IRs, which can affect integration efficiency and site recognition.
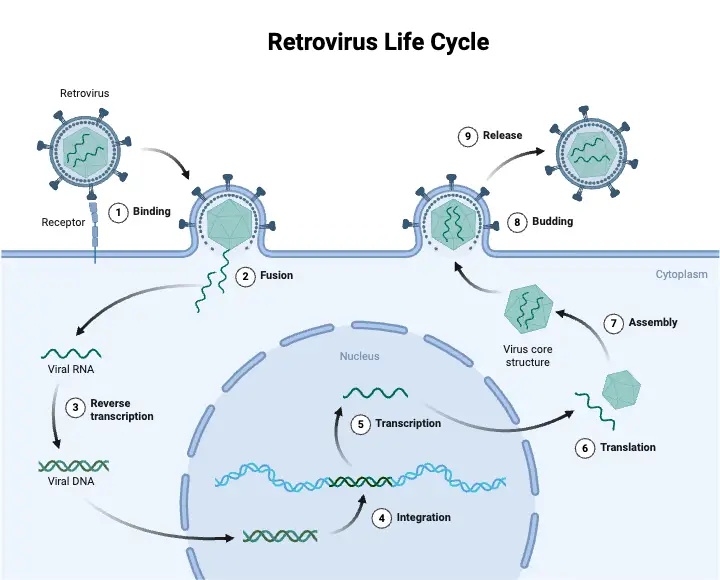
Retroviral Replication Cycle
The replication cycle of retroviruses is a complex process involving several key stages. Each step is crucial for the production of new viral particles and involves a series of well-coordinated events within the host cell.
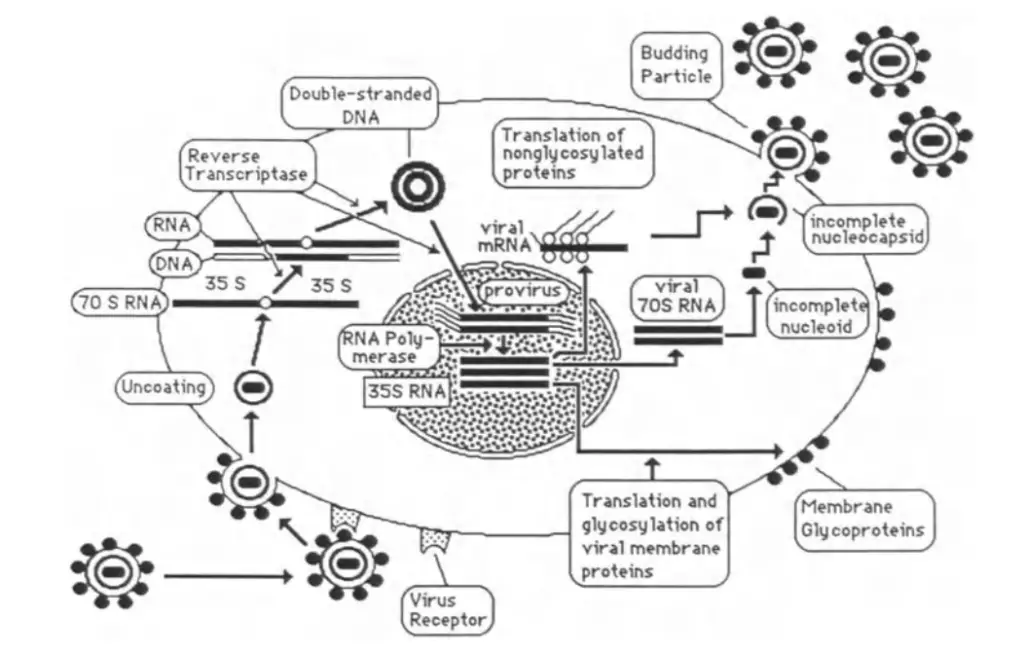
Step 1: Adsorption and Entry
- Adsorption to Host Cells:
- Retroviruses initiate infection by binding their envelope glycoproteins to specific receptors on the surface of a host cell.
- This binding is highly specific; only cells with the appropriate receptors can be infected.
- Entry into the Cell:
- After adsorption, the virus enters the cell through endocytosis or direct fusion with the plasma membrane.
- Once inside, the virus is uncoated, meaning the viral capsid is removed, and the RNA genome is released into the cytoplasm.
Step 2: Synthesis of Viral DNA
- Reverse Transcription:
- The viral RNA genome is reverse transcribed into double-stranded DNA by the enzyme reverse transcriptase.
- This process occurs in the cytoplasm and involves several activities of reverse transcriptase:
- DNA Polymerase Activity: Synthesizes the complementary DNA strand using the RNA as a template.
- RNase H Activity: Degrades the RNA strand from RNA-DNA hybrids, allowing synthesis of the second DNA strand.
- DNA Endonuclease Activity: Facilitates integration of the viral DNA into the host genome.
- Formation of Provirus:
- The newly synthesized double-stranded DNA is referred to as the provirus.
- This DNA integrates into the host cell’s genome, with regulatory sequences (LTRs) present at both ends of the integrated DNA.
Step 3: Integration into Host DNA
- Integration Process:
- The proviral DNA is inserted into the host cell’s DNA with the help of the host cell’s machinery and viral endonuclease activity.
- Integration can occur at various locations within the host genome and integrates in the same orientation as the original viral DNA.
- Persistence:
- Once integrated, the provirus becomes a permanent part of the host cell’s genome, which is replicated with each cell division.
- This integration allows for lifelong infection, as the provirus is passed on to daughter cells.
Step 4: Expression of Proviral DNA
- Transcription:
- The integrated proviral DNA is transcribed into RNA by the host cell’s RNA polymerase II.
- The RNA transcripts include both genomic RNA, which will be packaged into new virions, and mRNA, which will be translated into viral proteins.
- Regulation:
- In some retroviruses, regulatory proteins enhance transcription of the provirus. These are known as trans-acting transcriptional activators.
- Splicing and Translation:
- Viral mRNA undergoes splicing to produce various proteins, including structural proteins (gag, pol, env).
- Gag and gag-pol proteins are translated from full-length mRNA, while env proteins are translated from a smaller, spliced mRNA.
Step 5: Assembly and Release
- Protein Synthesis and Assembly:
- Viral proteins are synthesized and then assembled into new virions.
- Gag proteins form the core of the virus, while env proteins are integrated into the viral envelope.
- Budding:
- New virions are released from the host cell by a process called budding.
- The viral envelope is acquired from the host cell membrane, and the core is assembled either at or near the plasma membrane.
- Maturation:
- Newly formed virions often undergo maturation, including proteolytic cleavage of polyproteins into functional proteins.
- This maturation process is essential for producing fully infectious virions.
Adsorption and Entry Mechanism of Retroviruses
The process of retrovirus infection involves several key steps, starting with the adsorption of the virus to a host cell and culminating in its entry into the cell. This process is crucial for initiating the viral replication cycle. The main stages are outlined below:
- Binding to Host Cell Receptors
- Initial Attachment: The infection begins when the viral envelope glycoprotein binds to specific receptors on the host cell membrane. This interaction is highly specific; cells without these receptors are typically resistant to infection.
- Receptor Variety: A single retrovirus may use multiple receptors, as its envelope may contain several epitopes capable of binding to different membrane molecules.
- Multiple Receptors: The presence of more than one receptor type can enhance the virus’s ability to infect various cell types.
- Receptor Specificity: The specific interaction between viral glycoproteins and host cell receptors determines the virus’s tropism, or its preference for particular cell types.
- Entry into the Host Cell
- Mechanisms of Entry: Once bound, the virus enters the host cell through either endocytosis or direct penetration of the plasma membrane.
- Endocytosis: In this process, the host cell membrane engulfs the virus, forming a vesicle that brings the virus into the cytoplasm.
- Direct Penetration: Alternatively, some retroviruses fuse directly with the plasma membrane, releasing their contents into the cytoplasm.
- Uncoating: Following entry, the virus undergoes uncoating, a process wherein the viral capsid is partially degraded in the cytoplasm. This step converts the virus into a form that allows the initiation of DNA synthesis.
- Uncoating Mechanism: The exact mechanisms of uncoating are not fully understood, but it involves the release of viral RNA and proteins necessary for subsequent replication.
- Mechanisms of Entry: Once bound, the virus enters the host cell through either endocytosis or direct penetration of the plasma membrane.
- Post-Entry Events
- Viral Conversion: After uncoating, the viral RNA is converted into DNA. This newly synthesized DNA will integrate into the host cell’s genome, establishing a provirus that persists through cell division.
- Integration: The integration of viral DNA into the host genome is a critical step that ensures the virus’s replication and persistence within the host cell.
- Viral Conversion: After uncoating, the viral RNA is converted into DNA. This newly synthesized DNA will integrate into the host cell’s genome, establishing a provirus that persists through cell division.
Synthesis of Viral DNA Mechanism
The synthesis of viral DNA from single-stranded RNA is a crucial step in the retrovirus replication cycle. This process involves several stages and utilizes multiple enzymatic activities. The key points are outlined below:
- Reverse Transcription
- Conversion to DNA: The first step in viral DNA synthesis is the conversion of single-stranded viral RNA into double-stranded DNA. This process occurs in the cytoplasm of the host cell.
- Role of Reverse Transcriptase: Reverse transcription is mediated by the enzyme reverse transcriptase, which is a multifunctional protein complex. It requires a divalent cation, either magnesium (Mg²⁺) or manganese (Mn²⁺), depending on the specific virus.
- Enzymatic Activities: Reverse transcriptase exhibits three essential enzymatic activities:
- DNA Polymerase Activity: This activity synthesizes a complementary DNA strand (minus strand DNA) using the viral RNA as a template.
- RNase H Activity: This activity degrades the RNA strand from RNA-DNA hybrids, facilitating the synthesis of the second DNA strand (positive strand).
- DNA Endonuclease Activity: This activity is necessary for integrating the double-stranded viral DNA into the host cell’s genome.
- Enzymatic Activities: Reverse transcriptase exhibits three essential enzymatic activities:
- Formation of Long Terminal Repeats (LTRs)
- LTRs Formation: During reverse transcription, the regulatory sequences flanking the viral RNA are duplicated, leading to the formation of Long Terminal Repeats (LTRs) at both ends of the viral DNA. These LTRs consist of U3, R, and U5 regions.
- Importance of LTRs: LTRs are crucial for the integration of viral DNA into the host genome and for regulating the transcription of viral genes.
- LTRs Formation: During reverse transcription, the regulatory sequences flanking the viral RNA are duplicated, leading to the formation of Long Terminal Repeats (LTRs) at both ends of the viral DNA. These LTRs consist of U3, R, and U5 regions.
- Integration of Viral DNA
- Integration into Host Genome: The double-stranded viral DNA must be integrated into the host cell’s genome for successful viral replication. This integration is facilitated by the DNA endonuclease activity of reverse transcriptase.
- Role of Integrase: In addition to reverse transcriptase, the integrase enzyme, often encoded by the pol gene, plays a key role in the integration process. For example, in HIV, the pol gene encodes a protease that is distinct from integrase but is also involved in the viral life cycle.
- Integration into Host Genome: The double-stranded viral DNA must be integrated into the host cell’s genome for successful viral replication. This integration is facilitated by the DNA endonuclease activity of reverse transcriptase.
Integration of Viral DNA into Host Cell DNA: Mechanism
The integration of viral DNA into host cell DNA is a critical step in the retrovirus replication cycle. This process involves several key stages and components:
- Formation of Provirus
- Double-Stranded DNA Creation: The viral RNA, once reverse-transcribed into double-stranded DNA, is referred to as a provirus. This DNA is linear and must be integrated into the host cell’s genome to complete the viral replication cycle.
- Role of LTRs: Long Terminal Repeats (LTRs) at both ends of the viral DNA are crucial for integration. They contain regulatory elements essential for the integration process.
- Requirements for Successful Integration
- Intact LTRs: Both the 5′ and 3′ LTRs must be intact for proper integration of the viral DNA.
- Endonuclease Activity: Reverse transcriptase, while primarily known for its role in reverse transcription, also has endonuclease activity that facilitates integration. This activity is essential for making the initial cuts in the host DNA.
- Cell-Encoded Functions: Certain host cell factors are required to assist in the integration process. These factors help in recognizing and binding to the viral DNA.
- Integration Process
- Orientation and Site: Integration occurs in the same orientation as the viral DNA, specifically 5′ LTR-gag-pol-env-LTR 3′. The provirus can integrate at various sites within the host cell genome. Integration is generally non-specific, occurring at multiple potential sites in the host DNA.
- Unintegrated Viral DNA: Some viral DNA molecules may remain in the nucleus in an unintegrated form. These can be linear or circular and may replicate independently of the host genome. The persistence of unintegrated DNA is often associated with cytopathic effects and can contribute to cell damage and death.
- Implications of Integration
- Proviral Persistence: Once integrated, the viral DNA becomes part of the host cell’s genomic information. It is passed on to daughter cells during cell division. This integration makes retroviral infections chronic and lifelong.
- Latent Proviruses: Many integrated proviruses may remain latent, not actively expressing viral proteins. These latent proviruses are difficult for the immune system to detect, contributing to the persistence of the infection.
Expression of Proviral DNA: Mechanism
The expression of proviral DNA in retroviruses follows a well-defined process involving transcription and translation of viral genes. This mechanism is analogous to that of host cell gene expression, with specific modifications unique to retroviruses.
- Transcription of Proviral DNA
- Role of Cellular RNA Polymerase: Viral RNA is synthesized by the host cell’s type II RNA polymerase. The proviral genome functions as a transcriptional unit, with Long Terminal Repeats (LTRs) providing necessary regulatory sequences.
- Initiation and Enhancement: Transcription begins at the 5′ end of the R sequences in the 5′ LTR. LTRs contain signals that initiate and enhance transcription, guiding RNA polymerase to the start site.
- Polyadenylation: The RNA transcript is polyadenylated at the 3′ end, near the right end of the R sequence in the 3′ LTR, a process occurring post-transcriptionally.
- Trans-Acting Transcriptional Activators
- Human Retroviruses Exception: In contrast to most retroviruses, human retroviruses such as HIV, BLV, and visna virus produce proteins that enhance transcription. These proteins, known as trans-acting transcriptional activators (e.g., Tat in HIV), interact with the LTRs to boost RNA synthesis.
- Cellular Factors: The integration site of the provirus within the host DNA and the cell’s physiological state influence transcription. For instance, lymphocyte activation can stimulate HIV expression.
- RNA Processing and Protein Synthesis
- Primary RNA Transcript: The initial product of transcription is a full-length RNA molecule. This RNA can be used in two primary ways:
- Genomic RNA: About half of these transcripts serve as genomic RNA, incorporated into new virions.
- mRNA for Protein Production: The remaining transcripts undergo splicing to produce mRNA for viral protein synthesis.
- Splicing and Translation:
- Env Gene Products: Proteins encoded by the env gene are translated from a 4.0-kb spliced mRNA.
- Gag and Gag-Pol Proteins: These are synthesized from genome-sized mRNA, which includes overlapping open reading frames (ORFs) that utilize different reading frames.
- Protein Processing: Viral genes are often arranged in overlapping reading frames, allowing multiple proteins to be encoded from a single mRNA. The Gag, Pol, and Env proteins are initially synthesized as polyprotein precursors and later cleaved into functional proteins.
- Primary RNA Transcript: The initial product of transcription is a full-length RNA molecule. This RNA can be used in two primary ways:
- Quantitative Aspects of RNA and Protein Production
- RNA Abundance: Viral RNA constitutes a small fraction (0.1%-1%) of the total cellular RNA. It can represent up to 20% of the polyadenylated RNA in infected cells.
- Relative Protein Levels: Typically, more gag mRNA is synthesized compared to env mRNA, and gag levels are higher than pol mRNA.
Synthesis of Viral Proteins and Assembly of Virions: Mechanism
The synthesis of viral proteins and the assembly of virions involve intricate processes that ensure the creation of fully functional and infectious virus particles. This section details these mechanisms in a structured and comprehensive manner.
- Synthesis of Viral Proteins
- mRNA Splicing and Translation:
- Viral mRNA Splicing: After transcription, viral mRNA undergoes splicing to produce various proteins. This process is crucial for producing functional viral proteins.
- Gag and Pol Proteins: Gag proteins are synthesized as polyprotein precursors and later cleaved into multiple functional proteins. The Pol proteins are initially produced as large precursors, which are then processed into reverse transcriptase, integrase, and protease.
- Cleavage and Processing:
- Cleavage of Polyproteins: The primary Gag polyprotein is cleaved by a virus-encoded protease. This cleavage is often completed after the virion has budded from the host cell, resulting in immature particles.
- Processing of Env Proteins: The Env gene is translated into a large polyprotein precursor. This precursor is cleaved to produce surface glycoproteins essential for the virus’s ability to bind to host cells.
- mRNA Splicing and Translation:
- Assembly of Virions
- Core Assembly:
- Gag Proteins: Gag proteins self-assemble to form the viral core. This assembly occurs either at or near the plasma membrane or in the cytoplasm, depending on the virus type. The Gag proteins encapsidate the viral RNA, forming a core particle.
- Polyprotein Cleavage: The Gag polyproteins are cleaved into structural proteins that organize into a core structure. This process ensures the proper formation and maturation of the viral core.
- Envelope Formation:
- Budding Process: The envelope is derived from the host cell’s plasma membrane. During budding, the viral core acquires an envelope composed of host cell lipids and viral proteins.
- Interaction with Membrane: The viral envelope proteins, including Env, associate with the lipid membrane, facilitating the budding process. The N-terminal portion of Gag proteins may interact with the viral RNA and the envelope proteins.
- Types of Viruses and Assembly Differences:
- Type C Viruses: For type C retroviruses, core assembly occurs concurrently with budding. The Gag proteins and the viral RNA are packaged together as the virus buds from the cell membrane.
- Type B and D Viruses: These types may assemble their cores in the cytoplasm before interacting with the envelope. This distinction affects the timing and site of core assembly.
- Completion and Maturation:
- Immature Virions: Newly released virions often appear immature, containing uncleaved precursors. The maturation process involves further cleavage of polyproteins, which is critical for the production of infectious particles.
- Function of Protease: The viral protease, which is encoded by the pol gene, plays a key role in processing polyproteins. It cleaves the Gag and Pol precursors into functional viral proteins, ensuring the production of mature, infectious virions.
- Core Assembly:
Mechanisms of Retroviral Pathogenesis
- Neoplasia Induction
- Acute-Transforming Retroviruses:
- These viruses contain additional sequences known as oncogenes, which originate from cellular genes.
- Oncogenes promote rapid tumor formation by inducing inappropriate cellular growth. The viruses carrying these oncogenes are typically replication-defective, requiring coinfection with helper viruses for replication.
- Chronic-Transforming Retroviruses:
- These viruses lack cellular oncogenes but are replication-competent.
- They induce transformation through promoter insertion or insertional mutagenesis. Integration of the viral LTR near cellular proto-oncogenes results in their over-expression or dysregulated expression, leading to cancer. This process often has a prolonged latent period before disease onset.
- Acute-Transforming Retroviruses:
- Non-Oncogenic Retroviruses
- HTLV-I and HTLV-II:
- These viruses do not carry classical oncogenes and do not integrate at specific genome sites.
- They contain additional genes such as tat (transactivator of transcription) that interact with viral LTRs to enhance viral gene expression. These viruses can cause cellular transformation through mechanisms other than direct oncogene introduction.
- Bovine Leukosis Virus (BLV):
- Similar to HTLVs, BLV does not have classical oncogenes but contains extra coding sequences that contribute to its pathogenicity.
- HTLV-I and HTLV-II:
- HIV Pathogenesis
- Immunodeficiency:
- HIV primarily causes immunodeficiency by targeting and depleting helper T-cells.
- The virus produces several regulatory proteins, including tat, which facilitates transcriptional activation but differs from tat proteins in other retroviruses.
- HIV’s envelope proteins and interaction with the CD4 molecule contribute to the virus’s cytopathic effects. Additionally, monocyte/macrophage cells serve as reservoirs for the virus, influencing the disease’s progression.
- Immunodeficiency:
Clinical Syndromes of Retrovirus
Retroviruses are associated with various clinical syndromes in both humans and animals. These syndromes differ significantly based on the host species and the specific retrovirus involved. Here is a detailed overview:
Human Retroviruses
- Human T-Lymphotropic Viruses (HTLVs)
- Associated Conditions: HTLVs are primarily linked to T-cell lymphoma and leukemia.
- Mechanism: These viruses integrate their genetic material into T-cells, leading to malignancies. The integration can disrupt normal cellular regulation, resulting in uncontrolled T-cell proliferation.
Animal Retroviruses
- Sarcoma and Acute Leukemia Viruses
- Species Affected: These viruses predominantly affect animals and are not known to cause infections in humans.
- Effects: They induce conditions such as sarcoma and acute leukemia in various animal species.
- Examples: The avian leukosis complex virus is known to cause leukosis or sarcoma in fowls.
- Leukemia Viruses
- Characteristics: These are slow oncogenic viruses that induce malignancies after a long latency period, often up to 30 years.
- Mechanism: They promote cancer by indirectly influencing cellular growth. They contain a tax gene that acts as a transcriptional regulator, enhancing the expression of growth-promoting genes.
- Impact: This leads to prolonged periods of uncontrolled cell growth and potential neoplastic transformation.
- Murine Leukosis Viruses
- Associated Conditions: These include several strains linked to leukemia and sarcoma in mice.
- Special Case: The mammary tumor virus of mice is specifically associated with breast cancer in certain genetically predisposed mouse strains.
- Other Animal Retroviruses
- Species Affected: These viruses can induce leukosis and sarcoma in a range of animals, including cats, hamsters, rats, guinea pigs, and monkeys.
- Effects: The specific symptoms and severity can vary depending on the host and the virus strain.
Epidemiology of Retrovirus
Retroviruses exhibit a diverse epidemiology across different host species and geographical locations. Their transmission and host specificity are critical aspects of their spread and impact. Here is an in-depth look at the epidemiology of retroviruses:
Host Specificity and Transmission
- Host Specificity
- Viral Receptors: Retroviruses are highly specific to their host species. This specificity is determined by the presence of viral receptors on the host cell surfaces. The interaction between viral envelope proteins and these receptors dictates the ability of the virus to infect and replicate within a host.
- Cross-Species Transmission: While retroviruses are generally host-specific, some may cross species barriers under certain conditions, potentially leading to new infections in previously unexposed species.
- Transmission Pathways
- Exogenous Retroviruses
- Definition: These retroviruses are acquired from external sources.
- Transmission Mode: They are transmitted horizontally, meaning from one individual to another within the same species.
- Examples: Oncogenic retroviruses, which can induce cancer, are often exogenous. They spread through direct contact, contaminated surfaces, or body fluids.
- Endogenous Retroviruses
- Definition: These are inherited retroviruses, integrated into the host’s germ line DNA.
- Transmission Mode: They are transmitted vertically, from parents to offspring. This transmission occurs via the provirus integrated into the germ line cells.
- Characteristics: The provirus behaves like a cellular gene and is regulated by the host cell’s genetic machinery. Therefore, it is inherited with the host’s genetic material.
- Exogenous Retroviruses
Global Distribution and Impact
- Geographical Variation
- Worldwide Presence: Retroviruses are found in various host species across the globe. Their distribution is influenced by the host’s geographical range and ecological factors.
- Regional Differences: Some retroviruses may be more prevalent in certain regions due to local environmental conditions, host population density, and interaction with other species.
- Impact on Host Populations
- Human Health: Retroviruses such as HIV (Human Immunodeficiency Virus) and HTLV (Human T-Lymphotropic Virus) have significant impacts on human health, contributing to serious diseases like AIDS and adult T-cell leukemia.
- Animal Health: In animals, retroviruses can cause diseases such as leukemia and sarcoma. These diseases affect various species, including livestock and pets, impacting agricultural and veterinary sectors.
FAQ
What enzyme allows a retrovirus to make dna from an rna template?
The enzyme responsible for converting RNA into DNA in retroviruses is reverse transcriptase. This enzyme catalyzes the synthesis of complementary DNA (cDNA) from the viral RNA template. Reverse transcriptase has two main activities:
RNA-dependent DNA Polymerase Activity: Synthesizes DNA using the RNA genome as a template.
Ribonuclease H Activity: Degrades the RNA strand of the RNA-DNA hybrid, allowing for the synthesis of the second DNA strand.
These functions are crucial for the retroviral replication cycle, enabling the integration of viral DNA into the host genome.
how does a retrovirus like hiv enter a host cell?
A retrovirus like HIV enters a host cell through a well-coordinated process involving several key steps:
1. Attachment:
Viral Envelope Binding: HIV’s envelope protein, gp120, binds to the CD4 receptor on the surface of the host cell. CD4 is primarily found on T-helper cells.
Co-receptor Interaction: Following initial binding, gp120 also interacts with a co-receptor, typically CCR5 or CXCR4, which are additional surface proteins necessary for viral entry.
2. Fusion:
Conformational Change: The binding of gp120 to CD4 and the co-receptor induces a conformational change in the viral envelope protein.
Fusion Peptide Exposure: This change exposes the fusion peptide of another viral protein, gp41, which then interacts with the host cell membrane.
Membrane Fusion: gp41 facilitates the fusion of the viral envelope with the host cell membrane, allowing the viral core to enter the cell.
3. Entry and Uncoating:
Capsid Release: Once inside the host cell, the viral core, or capsid, disassembles, releasing the viral RNA and associated proteins into the cytoplasm.
4. Reverse Transcription:
Viral RNA to DNA: The released viral RNA is then reverse transcribed into DNA by the enzyme reverse transcriptase.
These steps enable HIV to establish infection and begin its replication cycle within the host cell.
References
- Schüpbach, J. (1990). Classification, Structure, and Biology of Retroviruses. In: Human Retrovirology. Current Topics in Microbiology and Immunology, vol 142. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-75195-0_2
- Coffin, J.M. (1992). Structure and Classification of Retroviruses. In: Levy, J.A. (eds) The Retroviridae. The Viruses. Springer, Boston, MA. https://doi.org/10.1007/978-1-4615-3372-6_2
- Voisset, Cécile & Weiss, Robin & Griffiths, David. (2008). Human RNA “Rumor” Viruses: the Search for Novel Human Retroviruses in Chronic Disease. Microbiology and molecular biology reviews : MMBR. 72. 157-96, table of contents. 10.1128/MMBR.00033-07.
- Petropoulos C. Retroviral Taxonomy, Protein Structures, Sequences, and Genetic Maps. In: Coffin JM, Hughes SH, Varmus HE, editors. Retroviruses. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1997. Available from: https://www.ncbi.nlm.nih.gov/books/NBK19417/
- Cloyd MW. Human Retroviruses. In: Baron S, editor. Medical Microbiology. 4th edition. Galveston (TX): University of Texas Medical Branch at Galveston; 1996. Chapter 62. Available from: https://www.ncbi.nlm.nih.gov/books/NBK7934/
- Saxena, S. K., & Chitti, S. V. (2016). Molecular Biology and Pathogenesis of Retroviruses. InTech. doi: 10.5772/62885