What is Restriction Fragment Length Polymorphism (RFLP)?
- Restriction Fragment Length Polymorphism (RFLP) is a valuable technique in molecular biology that allows for the differentiation and analysis of organisms based on the patterns derived from the cleavage of their DNA. This technique takes advantage of the natural variations present in homologous DNA sequences.
- At the core of RFLP is the existence of alternative alleles associated with restriction fragments that differ in size from one another. These variations in the length of restriction DNA fragments between individuals of a species are what define RFLP. To identify such polymorphisms, the DNA sample is first fragmented using a restriction enzyme. This enzyme recognizes and cuts DNA at specific short sequences, resulting in the formation of smaller fragments in a process called a restriction digest.
- The next step involves separating these DNA fragments by their length through agarose gel electrophoresis. This technique allows for the visualization and characterization of the fragments based on their size. The separated fragments are then transferred to a membrane using the Southern blot procedure. To determine the length of the fragments that are complementary to a specific DNA probe, the membrane is hybridized with the labeled probe.
- The detection of RFLPs occurs when the length of a specific fragment varies between individuals. These variations can provide valuable information for genetic studies and applications. However, it is worth noting that RFLP analysis has become less commonly used due to the advent of inexpensive DNA sequencing technologies that offer more comprehensive and efficient approaches.
- Despite its decreasing prevalence, RFLP analysis played a significant role in the early days of DNA profiling. It was the first DNA profiling technique that became widely accessible due to its relatively low cost. RFLP analysis was instrumental in genome mapping, localization of genes responsible for genetic disorders, determination of disease risk, and paternity testing. It provided valuable insights into the genetic makeup and relationships between individuals and populations.
- In summary, Restriction Fragment Length Polymorphism (RFLP) is a technique that exploits variations in homologous DNA sequences to differentiate organisms. By fragmenting DNA, separating the resulting fragments by size, and hybridizing them with labeled probes, RFLP analysis allows for the detection of polymorphisms. Although less commonly used now, RFLP analysis had a significant impact on genetic research and applications, particularly in the field of DNA profiling.
Principle of Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP) is a powerful technique used in molecular biology to analyze genetic variations and identify differences in DNA sequences. This technique relies on the principle that the DNA fragments generated by the digestion of DNA with specific restriction enzymes will differ in length based on the variations in the distance between the restriction sites.
In RFLP analysis, DNA samples are extracted from different organisms or individuals and then subjected to digestion with restriction enzymes. These enzymes recognize specific DNA sequences and cleave the DNA at those sites. If two organisms or individuals have variations in the distance between the recognition sites of a particular restriction enzyme, the resulting DNA fragments will differ in length.
The digested DNA fragments are then separated using a technique called gel electrophoresis. In this process, the DNA fragments are loaded onto an agarose gel and subjected to an electric current. The DNA fragments migrate through the gel based on their size, with smaller fragments traveling faster and longer fragments moving slower.
After the gel electrophoresis, the DNA fragments are visualized using staining techniques such as ethidium bromide. The resulting pattern of DNA bands on the gel represents the restriction fragment length polymorphism. By comparing the patterns obtained from different samples, scientists can identify similarities and differences in the DNA sequences.
The RFLP technique can be particularly useful in differentiating species and even strains from one another. Organisms that are closely related may have similar restriction fragment patterns, while those that are more distantly related may display distinct patterns. By analyzing the RFLP patterns, scientists can gain insights into the genetic diversity and relatedness among different organisms.
In addition to species differentiation, RFLP analysis has been extensively used in various fields such as forensic science, genetic mapping, and disease diagnosis. For example, in forensic investigations, RFLP can be employed to match DNA samples found at a crime scene with those of potential suspects. In genetic mapping studies, RFLP markers can be used to identify the location of disease-related genes on chromosomes.
In conclusion, the principle of Restriction Fragment Length Polymorphism (RFLP) relies on the variations in the length of DNA fragments generated by the digestion of DNA with specific restriction enzymes. By analyzing the patterns of these fragments using gel electrophoresis, scientists can differentiate species and strains, assess genetic diversity, and gain insights into various genetic phenomena. RFLP continues to be a valuable tool in molecular biology research, enabling the exploration of genetic variations and their implications in various fields.
Steps of Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP) is a technique used to analyze genetic variations in DNA sequences. The following steps outline the process of RFLP analysis:
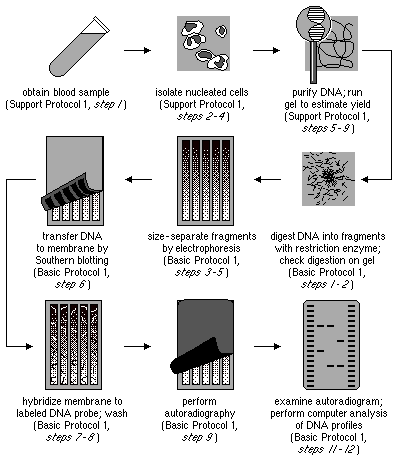
- DNA Isolation: The first step is to isolate the DNA from the target organism or sample. This involves extracting DNA from cells or tissues using various methods, ensuring the DNA is of high quality and purity.
- Restriction Digestion: Once the DNA is isolated, it is subjected to restriction digestion using specific restriction enzymes. These enzymes recognize and cleave DNA at specific recognition sites. The DNA is incubated with the restriction enzymes, resulting in the fragmentation of DNA into smaller fragments.
- Gel Electrophoresis: The digested DNA sample is then separated using gel electrophoresis. In this process, the DNA fragments are loaded onto an agarose gel and subjected to an electric current. The DNA fragments migrate through the gel based on their size, with smaller fragments moving faster and longer fragments moving slower. This separation allows for the visualization and analysis of the DNA fragments.
- Denaturation: After gel electrophoresis, the gel is exposed to a denaturing solution, which denatures the double-stranded DNA fragments, converting them into single-stranded DNA. This step is essential for further analysis.
- Southern Blotting: Following denaturation, the DNA fragments on the gel are transferred onto a nylon membrane through a technique called Southern blotting. The nylon membrane allows for the immobilization of the DNA fragments while preserving their spatial arrangement from the gel.
- Probe Hybridization: The nylon membrane containing the transferred DNA fragments is then exposed to a solution containing complementary nucleotide probes. These probes are typically labeled with radioactive or fluorescent tags and are designed to specifically hybridize with chosen DNA sequences on the nylon membrane. The hybridization occurs between the probes and the complementary DNA sequences on the membrane.
- Autoradiography: After probe hybridization, the membrane is placed against X-ray film. The hybridized radioactive probes on the membrane cause exposure of the X-ray film, producing an autoradiogram. The autoradiogram represents the pattern of DNA fragments and serves as a visual record of the hybridized DNA sequences.
- RFLP Analysis: The final step involves the analysis of the autoradiogram to detect and compare differences in DNA fragment patterns among samples. Polymorphisms, which are variations in the DNA sequence that result in different fragment patterns, can be identified. By comparing the patterns, scientists can differentiate between individuals or species, assess genetic diversity, and gain insights into genetic relationships.
RFLP analysis
Restriction Fragment Length Polymorphism (RFLP) analysis is a technique used to detect genetic variation by examining the lengths of DNA fragments produced by the digestion of DNA samples with specific restriction enzymes. RFLP analysis has been widely used in genetic research, forensic investigations, and paternity testing. Here’s how RFLP analysis works:
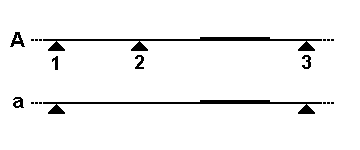
- Restriction Digestion: The first step in RFLP analysis is the fragmentation of DNA using restriction enzymes. These enzymes recognize and cleave DNA at specific sequences, generating DNA fragments of varying lengths. The selection of the appropriate restriction enzyme is crucial for targeting specific regions of the DNA.
- Gel Electrophoresis: After restriction digestion, the DNA fragments are separated based on their size using a technique called agarose gel electrophoresis. The DNA fragments are loaded onto a gel and subjected to an electric field. Smaller fragments move more quickly through the gel matrix, while larger fragments move more slowly. As a result, the DNA fragments separate into distinct bands based on their size.
- Southern Blotting: The separated DNA fragments are then transferred from the gel onto a membrane in a process called Southern blotting. The membrane, typically made of nylon, allows for the immobilization of the DNA fragments while preserving their spatial arrangement from the gel.
- Hybridization: The membrane with the immobilized DNA fragments is then exposed to a labeled DNA probe that is complementary to a specific sequence of interest. The probe binds to its complementary sequence on the membrane, forming a hybridization. The probe is usually labeled with a radioactive or fluorescent marker, allowing for the detection of the hybridized fragments.
- Detection and Analysis: After hybridization, the membrane is visualized to identify the DNA fragments that have hybridized with the probe. The length of the detected fragments indicates the presence or absence of specific DNA sequences or variations. Variations in fragment lengths between individuals indicate the occurrence of a restriction fragment length polymorphism.
RFLP analysis can detect genetic variations resulting from sequence differences, insertions, deletions, translocations, inversions, and other genetic processes. These variations can be used as genetic markers for subsequent genetic analysis. RFLP analysis requires larger amounts of DNA compared to other techniques like short tandem repeat (STR) analysis.
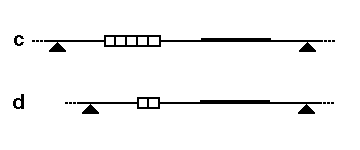
In conclusion, RFLP analysis is a powerful tool for detecting genetic variation. By analyzing the lengths of DNA fragments generated through restriction digestion and gel electrophoresis, RFLP analysis enables the identification of restriction fragment length polymorphisms. These polymorphisms serve as genetic markers and can be utilized in diverse fields such as genetic research, forensic analysis, and paternity testing.
Applications of Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP) analysis has found diverse applications in various fields of research and practical use. The following are some key applications of RFLP:
- Forensic Analysis: RFLP is commonly used in forensic investigations to determine the source of DNA samples. By comparing RFLP patterns between crime scene DNA and potential suspects’ DNA, forensic scientists can identify matches or exclusions, aiding in the resolution of criminal cases.
- Genetic Disease Detection: RFLP analysis can be employed to determine the disease status of an individual. By identifying specific RFLP patterns associated with genetic mutations or disease-related genes, researchers can assess an individual’s risk for certain disorders or predict the presence of genetic abnormalities.
- Genetic Mapping: RFLP has been instrumental in genome mapping and the localization of genes responsible for genetic disorders. By measuring recombination rates between RFLP loci, researchers can create genetic maps that provide insights into the arrangement and distances between genes. This information is crucial for understanding the genetic basis of diseases and facilitating further research in the field of genetics.
- Population Genetics and Breeding Studies: RFLP analysis has been widely used in the characterization of genetic diversity and breeding patterns in animal populations. By examining the RFLP patterns within a population, researchers can assess genetic variations, identify individuals with unique or rare genetic markers, and study breeding patterns and population structure. This information is valuable in conservation biology, animal breeding programs, and evolutionary studies.
- Chromosome Mapping: RFLP analysis has been successfully applied to chromosome mapping in various organisms, including humans, mice, maize, tomato, rice, and more. By mapping RFLP markers onto chromosomes, researchers can determine the positions of genes and genetic loci of interest. This information aids in understanding the organization and structure of chromosomes, identifying gene linkages, and studying genetic traits and inheritance patterns.
Advantages of Restriction Fragment Length Polymorphism (RFLP)
Restriction Fragment Length Polymorphism (RFLP) analysis offers several advantages that contribute to its wide application in genetic research. The following are key advantages of RFLP:
- No Prior Sequence Information Required: One of the primary advantages of RFLP analysis is that it does not require prior knowledge of DNA sequence information or the synthesis of specific oligonucleotides. Unlike PCR-based protocols that rely on primer design and sequence specificity, RFLP analysis can be performed without the need for sequence data. This makes it a valuable tool when working with organisms for which sequence information may be limited or unavailable.
- Genotypic Characteristics: RFLP analysis is based on genotypic characteristics rather than phenotypic traits. This means that RFLP patterns are determined by the underlying genetic variations in DNA sequences, allowing for accurate and reliable identification of genetic markers. By focusing on genotypic information, RFLP analysis provides insights into the genetic composition and diversity of individuals or populations.
- Co-Dominant Marker: RFLP is considered a co-dominant genetic marker. This means that heterozygosity, or the presence of different alleles at a given locus, can be accurately estimated using RFLP analysis. The co-dominant nature of RFLP allows for a more comprehensive understanding of genetic variation within a population.
- Useful in Genomic DNA Sequencing: RFLP analysis is highly useful in studying genomic DNA sequences. It provides a means to identify specific DNA variations, such as single nucleotide polymorphisms (SNPs) or larger insertions and deletions. By detecting and analyzing these variations, RFLP analysis contributes to understanding the structure, organization, and genetic traits associated with genomic DNA sequences.
- Robust Methodology: RFLP analysis is a highly robust methodology that can be easily standardized and has good transferability between laboratories. The protocol and techniques involved in RFLP analysis have been well-established and widely used, ensuring consistent and reliable results. The robust nature of RFLP analysis makes it a preferred choice for various genetic research applications.
Limitations of Restriction Fragment Length Polymorphism (RFLP)
While Restriction Fragment Length Polymorphism (RFLP) analysis has been widely used in genetic research, it also has certain limitations that should be considered. The following are key limitations of RFLP:
- Slow Process: RFLP analysis can be a time-consuming process. It involves several steps, including DNA extraction, restriction digestion, gel electrophoresis, hybridization, and autoradiography. Each step requires careful optimization and can contribute to a longer turnaround time compared to some other genetic analysis methods.
- Cumbersome Methodology: The process of RFLP analysis can be cumbersome and labor-intensive. It involves multiple manual steps, such as probe labeling, DNA fragmentation, gel loading, blotting, hybridization, washing, and autoradiography. This complexity increases the chances of errors and requires careful attention and expertise.
- Large Amount of Sample DNA: RFLP analysis typically requires a substantial amount of DNA sample for accurate analysis. The restriction digestion and subsequent electrophoresis and blotting processes necessitate a sufficient quantity of DNA for reliable results. This can pose challenges when working with limited or precious DNA samples, such as in ancient DNA or forensic investigations.
- Lack of Automation: Unlike some newer genetic analysis techniques, RFLP analysis is not easily automated. The manual nature of the process, including gel electrophoresis and autoradiography, makes it difficult to streamline and automate the workflow. This can limit its scalability and efficiency, especially when dealing with a large number of samples.
- Low Polymorphism in Some Species: RFLP analysis relies on detecting genetic variations and polymorphisms within DNA sequences. However, in certain species or populations, the level of genetic polymorphism may be low, resulting in limited or less informative RFLP patterns. This can reduce the effectiveness and discriminatory power of RFLP analysis in such cases.
- Limited Loci Detected: Each RFLP assay typically detects only a limited number of loci or regions of interest. The number of DNA fragments that can be resolved on a gel and analyzed simultaneously is constrained, restricting the scope of analysis within a single assay. This limitation may require multiple assays or the use of additional genetic markers to obtain a more comprehensive understanding of the genetic landscape.
- Requirement for Suitable Probe Library: RFLP analysis relies on the availability of suitable probe libraries containing complementary nucleotide sequences for hybridization. The generation and maintenance of these libraries require resources, time, and expertise. The lack of an appropriate probe library can limit the application of RFLP analysis.
FAQ
What is Restriction Fragment Length Polymorphism (RFLP)?
Restriction Fragment Length Polymorphism (RFLP) is a technique used to detect genetic variations by analyzing the lengths of DNA fragments produced through the digestion of DNA with specific restriction enzymes.
What are the applications of RFLP analysis?
RFLP analysis has applications in genome mapping, localization of genes for genetic disorders, disease risk determination, paternity testing, forensic investigations, measuring recombination rates, and studying genetic diversity in populations.
What are the advantages of RFLP analysis?
The advantages of RFLP analysis include not requiring prior sequence information, reliance on genotypic characteristics, estimation of heterozygosity, usefulness in studying genomic DNA sequences, and a robust and transferable methodology.
What are the limitations of RFLP analysis?
RFLP analysis has limitations such as being slow and cumbersome, requiring a large amount of sample DNA, lack of automation, low polymorphism in some species, detection of few loci per assay, and the need for a suitable probe library.
How does RFLP differ from PCR?
RFLP analysis and PCR (Polymerase Chain Reaction) are both techniques used in genetic analysis, but they differ in their approaches. RFLP requires the use of restriction enzymes to digest DNA, while PCR amplifies specific DNA regions using primers and a DNA polymerase enzyme.
How does RFLP analysis work?
RFLP analysis involves fragmenting DNA using restriction enzymes, separating the fragments based on their size through gel electrophoresis, transferring them onto a membrane via Southern blotting, and hybridizing the membrane with a labeled DNA probe to identify and analyze the lengths of the fragments.
Can RFLP analysis detect single nucleotide polymorphisms (SNPs)?
RFLP analysis can indirectly detect single nucleotide polymorphisms (SNPs) if the SNP alters the recognition site for a restriction enzyme, leading to different fragment lengths. However, direct detection of SNPs is better achieved through other techniques like PCR-based methods or sequencing.
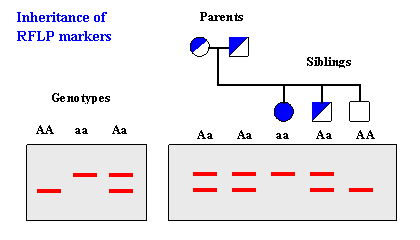
How is RFLP analysis helpful in paternity testing?
RFLP analysis can be used in paternity testing by comparing the DNA profiles of the child and potential parents. If there are differences in the RFLP patterns, it suggests a non-parental relationship, while matching patterns indicate a high probability of a biological relationship.
Can RFLP analysis be automated?
Unlike some newer genetic analysis techniques, RFLP analysis is not easily automated due to its manual steps such as gel electrophoresis and autoradiography. However, certain aspects of the process, such as probe labeling and hybridization, can be automated to some extent.
Are there alternative techniques to RFLP analysis?
Yes, several alternative techniques have been developed that have largely replaced RFLP analysis for certain applications. These include PCR-based methods like Short Tandem Repeat (STR) analysis, Single Nucleotide Polymorphism (SNP) genotyping, and Next-Generation Sequencing (NGS) technologies.
References
- Botstein, D., White, R.L., Skolnick, M., and Davis, R.W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32(3), 314-331.
- Kwok, S., and Higuchi, R. (1989). Avoiding false positives with PCR. Nature, 339(6221), 237-238.
- Saiki, R.K., Scharf, S., Faloona, F., Mullis, K.B., Horn, G.T., Erlich, H.A., and Arnheim, N. (1985). Enzymatic amplification of beta-globin genomic sequences and restriction site analysis for diagnosis of sickle cell anemia. Science, 230(4732), 1350-1354.
- Zhou, H.H., Jiang, Z.M., Zhao, Y.R., Wang, J.B., Zhang, Z.P., and Wang, H. (2009). Application of RFLP-PCR for paternity testing of Phasianus colchicus. Yi Chuan, 31(1), 98-104.
- Kumar, V., and Abbas, A.K. (2020). Robbins Basic Pathology. Elsevier Health Sciences.
- Lodish, H., Berk, A., Zipursky, S.L., Matsudaira, P., Baltimore, D., and Darnell, J. (2000). Molecular Cell Biology. W.H. Freeman and Company.