What is Protein Synthesis?
Protein synthesis is a fundamental biological process that occurs within cells, responsible for the formation of proteins through the assembly of amino acid chains, termed polypeptides. This process is pivotal to the central dogma of genetics and is instrumental in maintaining cellular function and structure.
The process of protein synthesis can be broadly categorized into two phases: transcription and translation.
- Transcription: During this phase, a specific segment of DNA, referred to as a gene, serves as a template for the synthesis of a molecule called messenger RNA (mRNA). This transcription is facilitated by enzymes known as RNA polymerases, which operate within the cell’s nucleus. In eukaryotic organisms, the initially formed mRNA, termed pre-mRNA, undergoes a series of post-transcriptional modifications to yield mature mRNA. Subsequently, this mature mRNA is transported from the nucleus to the cell’s cytoplasm through nuclear pores, setting the stage for the next phase.
- Translation: Here, the mRNA serves as a template that is interpreted by cellular structures called ribosomes. These ribosomes read the codon sequences in the mRNA and, with the assistance of transfer RNA (tRNA) molecules, ensure the correct sequence of amino acids is assembled. This process is sequential and can be divided into three stages: initiation, elongation, and termination. The outcome is a polypeptide chain, which then undergoes folding to adopt its functional three-dimensional shape. This folding process involves the formation of secondary structures, which then coalesce to form the protein’s tertiary structure. Once properly folded, proteins may undergo further modifications, termed post-translational modifications, which can influence their function, location within the cell, and interactions with other proteins.
Therefore, it is evident that protein synthesis is not merely the formation of polypeptide chains. It is a complex, multi-step process that involves various cellular components, from DNA and RNA to ribosomes and enzymes. Besides its role in producing functional proteins, this process also plays a crucial role in disease pathology. Errors in protein synthesis, stemming from DNA mutations or protein misfolding, can lead to diseases. For instance, DNA mutations can alter the mRNA sequence, leading to changes in the resultant protein’s amino acid sequence. Such changes can affect the protein’s function or its ability to fold correctly. Then, misfolded proteins, which often aggregate, have been implicated in numerous diseases, notably neurological disorders like Alzheimer’s and Parkinson’s disease.
In conclusion, protein synthesis is an intricate process that ensures the correct formation and functioning of proteins within cells. It involves a series of well-coordinated steps, from the transcription of DNA to the translation of mRNA, culminating in the formation of functional proteins. Any disruptions in this process can have significant implications for cellular function and health.
Protein synthesis definition
Protein synthesis is the cellular process in which proteins are produced by decoding the genetic information stored in DNA into amino acid sequences, involving two main stages: transcription (where DNA is transcribed into mRNA) and translation (where mRNA is translated into a polypeptide chain by ribosomes).
Site of protein synthesis – Where does protein synthesis take place?
The site of protein synthesis varies depending on the type of organism. In general, protein synthesis occurs in specific cellular compartments or structures. Here are the main sites of protein synthesis in different organisms:
- Prokaryotes (such as bacteria): Protein synthesis occurs in the cytoplasm of prokaryotic cells. Ribosomes, the cellular machinery responsible for protein synthesis, attach to the mRNA and carry out translation in the cytoplasmic environment.
- Eukaryotes (including animals, plants, and fungi): In eukaryotic cells, protein synthesis takes place in two main locations:
- a. Cytoplasm: The majority of protein synthesis occurs in the cytoplasm of eukaryotic cells. Cytoplasmic ribosomes (free ribosomes) are responsible for translating most mRNA molecules into proteins.
- b. Rough endoplasmic reticulum (ER): Some proteins are synthesized on ribosomes that are bound to the rough ER. These proteins are typically destined for secretion or integration into cellular membranes. The rough ER provides a site for co-translational processing, folding, and modification of the synthesized proteins. It’s important to note that protein synthesis starts in the cytoplasm regardless of the final destination of the protein. Many proteins, after being synthesized in the cytoplasm, may undergo further processing and modifications in specific organelles before reaching their functional locations.
- Organelles within eukaryotic cells: Certain organelles within eukaryotic cells also have their own specialized protein synthesis machinery:
- a. Mitochondria: Mitochondria, the cellular powerhouses responsible for energy production, have their own ribosomes and carry out protein synthesis for a subset of mitochondrial proteins.
- b. Chloroplasts: In plant cells, chloroplasts, the site of photosynthesis, also have their own ribosomes and conduct protein synthesis specifically for chloroplast proteins.
Protein Synthesis Machinery
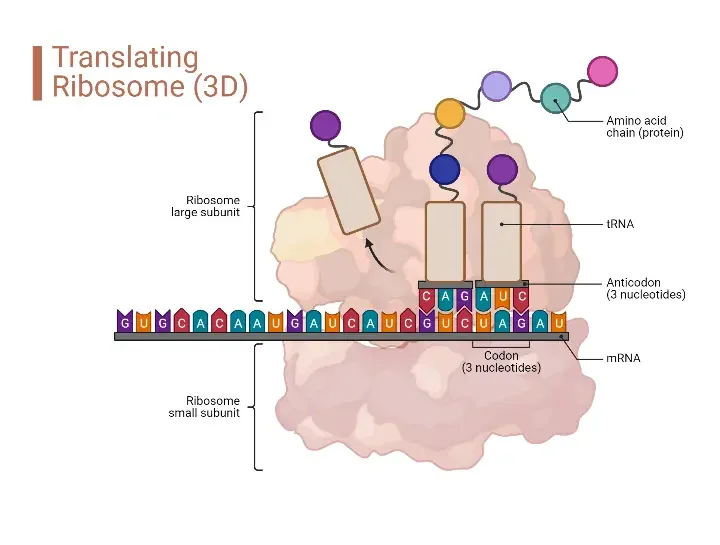
Protein synthesis, a pivotal cellular process, involves the decoding of genetic information to produce proteins. This intricate process requires a set of specialized components, collectively termed the protein synthesis machinery. The following sections provide a detailed overview of these components and their respective functions.
- Ribosomes Ribosomes are complex structures composed of ribosomal RNA (rRNA) and proteins. Often referred to as ribozymes due to the enzymatic activity of rRNA, they play a central role in protein synthesis. The ribosome consists of two subunits, each with specific active sites (E, P, A) crucial for its catalytic activity. These sites facilitate the sequential addition of amino acids during translation.
- Messenger RNA (mRNA) Serving as a template, mRNA carries the genetic instructions from DNA to the ribosomes. It encodes the sequence of amino acids required for the specific protein being synthesized. After being transcribed from DNA in the nucleus, mRNA molecules are transported to the cytoplasm, where they guide the translation process.
- Transfer RNA (tRNA) tRNA molecules are instrumental in translating the genetic code from mRNA into a sequence of amino acids. Each tRNA molecule is specific to a particular amino acid and carries an anticodon complementary to the codon on mRNA. For instance, if lysine is coded by the codon AAG, the corresponding tRNA will have the anticodon UUC. As tRNA binds to mRNA, it releases its associated amino acid, allowing the ribosome to form bonds between consecutive amino acids, thereby elongating the polypeptide chain.
- Amino Acids Amino acids, the fundamental building blocks of proteins, are attached to their respective tRNA molecules. These amino acids are then transported to the ribosome during translation. The sequence of codons on the mRNA determines the specific order in which these amino acids are assembled into a protein.
- Initiation Factors These are proteins essential for the onset of translation. Initiation factors aid in assembling the ribosome on the mRNA, ensuring it is correctly positioned at the start codon and maintaining the appropriate reading frame for protein synthesis.
- Elongation Factors As the name suggests, elongation factors assist during the elongation phase of protein synthesis. They ensure the accurate and efficient addition of amino acids to the growing polypeptide chain.
- Release Factors Recognizing the stop codon on mRNA, release factors signal the end of protein synthesis. They facilitate the detachment of the completed polypeptide chain from the ribosome, marking the conclusion of the translation process.
Protein Synthesis Steps/Steps of Protein synthesis
Protein synthesis is a complex cellular process that involves the creation of proteins from the instructions encoded in DNA. It occurs in several steps, which include transcription and translation.
- Transcription: The first step of protein synthesis is transcription, which takes place in the nucleus of eukaryotic cells. During transcription, the DNA sequence of a specific gene is copied into a molecule called messenger RNA (mRNA). This process involves the enzyme RNA polymerase, which binds to the DNA and synthesizes a complementary mRNA strand.
- mRNA Processing: After transcription, the newly synthesized mRNA undergoes processing before leaving the nucleus. This includes the addition of a protective cap at the 5′ end and a poly-A tail at the 3′ end. Additionally, any non-coding regions called introns are removed, and the remaining coding regions called exons are spliced together to form the mature mRNA molecule.
- Translation Initiation: The processed mRNA molecule exits the nucleus and travels to the cytoplasm, where translation occurs. The ribosomes, the cellular structures responsible for protein synthesis, attach to the mRNA at a specific region called the start codon. This process is known as translation initiation.
- Elongation: During the elongation phase, the ribosome moves along the mRNA molecule, reading the genetic code in sets of three nucleotides called codons. Each codon corresponds to a specific amino acid. Transfer RNA (tRNA) molecules bring the corresponding amino acids to the ribosome, and the ribosome connects them together in the order dictated by the mRNA codons. This forms a growing polypeptide chain.
- Termination: The termination phase marks the end of protein synthesis. When the ribosome reaches a stop codon on the mRNA, it recognizes that protein synthesis is complete. At this point, the newly synthesized polypeptide chain is released from the ribosome.
- Post-Translational Modifications: After synthesis, the polypeptide chain may undergo various post-translational modifications, such as folding into its proper three-dimensional structure, undergoing chemical modifications (e.g., addition of functional groups), or being targeted for specific destinations within the cell.
These steps of protein synthesis ensure the accurate translation of genetic information from DNA to functional proteins, which play crucial roles in cellular processes, organismal development, and overall biological functioning.
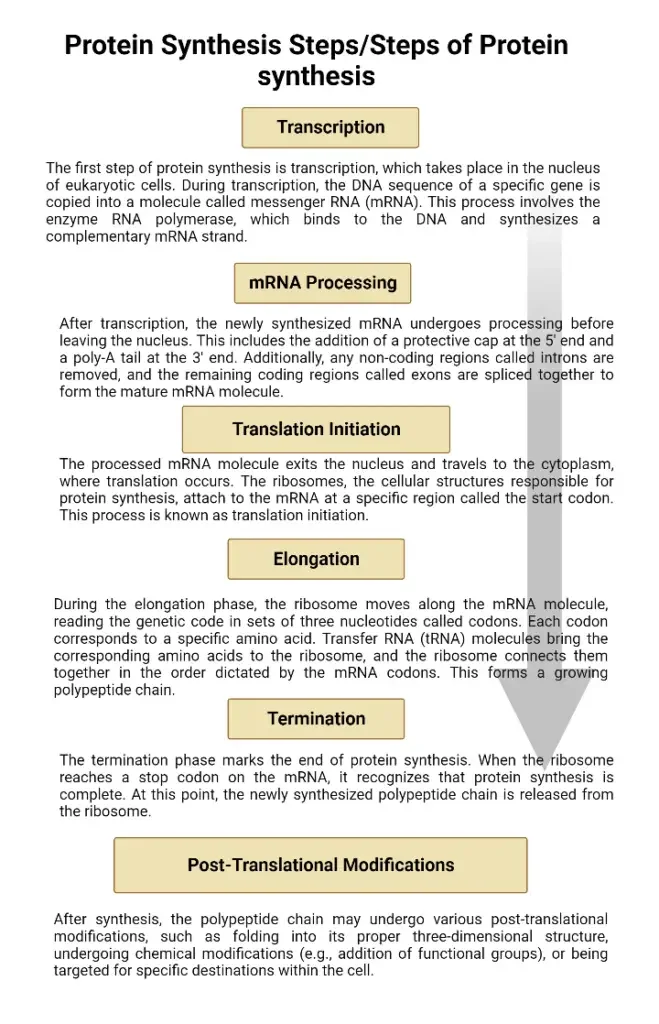
Labeled diagram of protein synthesis
Enzymes involve in Protein Synthesis and their functions
- RNA Polymerase RNA polymerase is instrumental in the transcription phase, where it synthesizes messenger RNA (mRNA) using a DNA template. By binding to specific regions of the DNA, it catalyzes the formation of an RNA strand that is complementary to the DNA sequence.
- DNA Helicase During transcription, DNA helicase unwinds the double-stranded DNA, creating what is termed the transcription bubble. This unwinding allows RNA polymerase to access the DNA template, facilitating RNA synthesis.
- RNA Processing Enzymes Post-transcription, the newly formed mRNA undergoes several modifications. Enzymes such as RNA splicing factors come into play, removing introns (non-coding regions) and splicing together exons (coding regions). This results in a mature mRNA molecule, ready for translation.
- Ribosomes While not enzymes in the traditional sense, ribosomes are essential components of the protein synthesis machinery. Comprising proteins and ribosomal RNA (rRNA), ribosomes provide a platform for mRNA and transfer RNA (tRNA) interaction during translation. This ensures the correct sequence of amino acids in the nascent protein.
- Aminoacyl-tRNA Synthetases These enzymes have a pivotal role in translation. They recognize specific amino acids and attach them to their corresponding tRNA molecules. This ensures that the correct amino acids are incorporated into the growing protein chain based on the mRNA template.
- Peptidyl Transferase Located within ribosomes, peptidyl transferase is a key enzyme in the translation process. Its primary function is to catalyze the formation of peptide bonds between adjacent amino acids, facilitating the growth of the polypeptide chain. This enzyme, unique in its composition, is entirely made up of RNA. Specifically, in prokaryotes, the 23S rRNA subunit contains the peptidyl transferase, while in eukaryotes, it resides in the 28S subunit. Therefore, its mechanism is mediated by ribosomal RNA (rRNA), functioning as a ribozyme.
- Release Factors Concluding the translation process are the release factors. These enzymes detect specific termination codons on the mRNA. Upon recognition, they promote the detachment of the completed protein from the ribosome, marking the end of protein synthesis.
| Enzyme/Component | Function |
|---|---|
| RNA Polymerase | Catalyzes the synthesis of messenger RNA (mRNA) using a DNA template during transcription. |
| DNA Helicase | Unwinds the double-stranded DNA during transcription, creating the transcription bubble, which allows RNA polymerase to access the DNA template. |
| RNA Processing Enzymes | Modify the newly formed mRNA post-transcription by removing introns and splicing together exons, resulting in a mature mRNA molecule. |
| Ribosomes | Comprising proteins and ribosomal RNA (rRNA), they provide a platform for mRNA and transfer RNA (tRNA) interaction during translation, ensuring the correct sequence of amino acids in the protein. |
| Aminoacyl-tRNA Synthetases | Recognize specific amino acids and attach them to their corresponding tRNA molecules, ensuring the correct amino acids are incorporated into the growing protein chain based on the mRNA template. |
| Peptidyl Transferase | Located within ribosomes, it catalyzes the formation of peptide bonds between adjacent amino acids during translation. Unique in its composition, it is entirely made up of RNA, with its mechanism mediated by ribosomal RNA (rRNA). |
| Release Factors | Detect specific termination codons on the mRNA during translation. Upon recognition, they promote the detachment of the completed protein from the ribosome, marking the end of protein synthesis. |
An Overview of the Protein Synthesis (Translation)
Protein synthesis is the cellular process responsible for translating genetic information into functional proteins. This intricate mechanism involves multiple stages and components, each playing a pivotal role in ensuring the accurate production of proteins.
- Initiation of Translation The process of protein synthesis commences with the initiation of ribosomal translation. During this phase, ribosomes identify and bind to the starting point on the mRNA. Concurrently, a tRNA molecule carrying a specific amino acid attaches itself to the mRNA. In prokaryotic organisms, this initial amino acid is N-formylmethionine.
- Elongation Following initiation, the elongation phase begins. Here, the second amino acid is linked to the first, facilitated by the ribosome. The ribosome then shifts its position along the mRNA, systematically reading the codons and ensuring the correct sequence of amino acids is added to the growing polypeptide chain. This elongation cycle is repeated until the ribosome encounters a stop codon on the mRNA.
- Protein Folding Once the elongation process reaches the stop codon, the newly formed amino acid chain undergoes spontaneous folding to adopt its functional three-dimensional structure, resulting in a protein. The precise folding of the protein is crucial for its functionality and activity within the cell.
- Termination and Ribosome Recycling Upon reaching the stop codon, the translation process concludes. Subsequently, the ribosome dissociates into its two constituent subunits. These subunits, though separated, can later reassociate in preparation for translating another mRNA molecule.
- Role of Catalytic Proteins and GTP The efficiency and accuracy of protein synthesis are enhanced by several catalytic proteins, including initiation, elongation, and termination factors. Besides these factors, guanosine triphosphates (GTP) play a significant role in the process. GTP molecules release energy upon conversion to guanosine diphosphate (GDP), providing the necessary energy for various stages of protein synthesis.
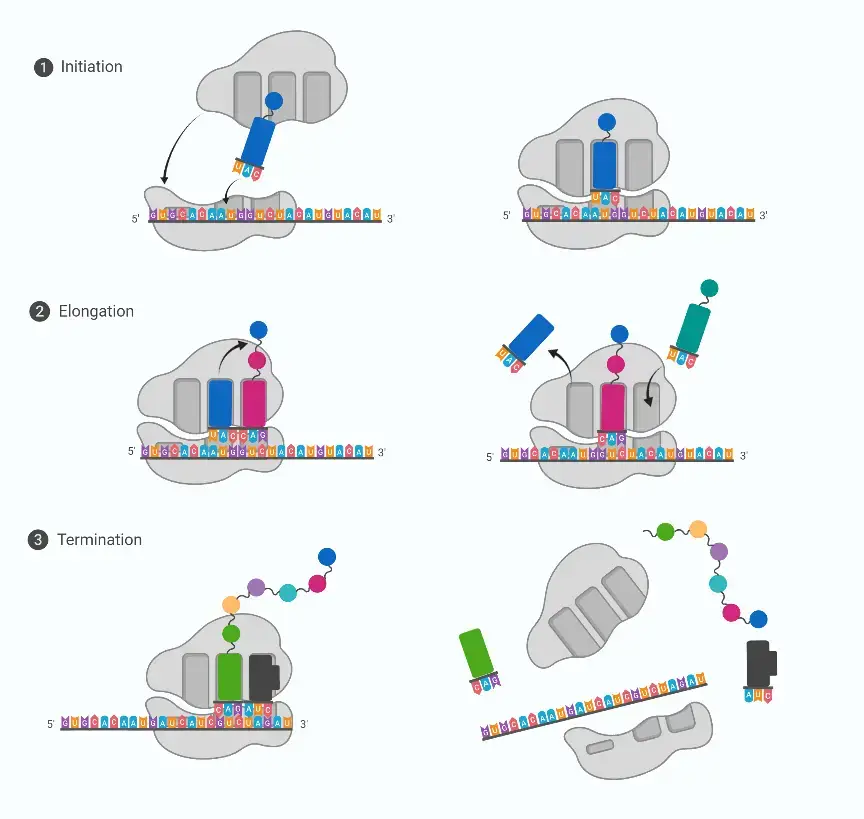
Process of Protein synthesis/Protein Synthesis Steps(Translation)
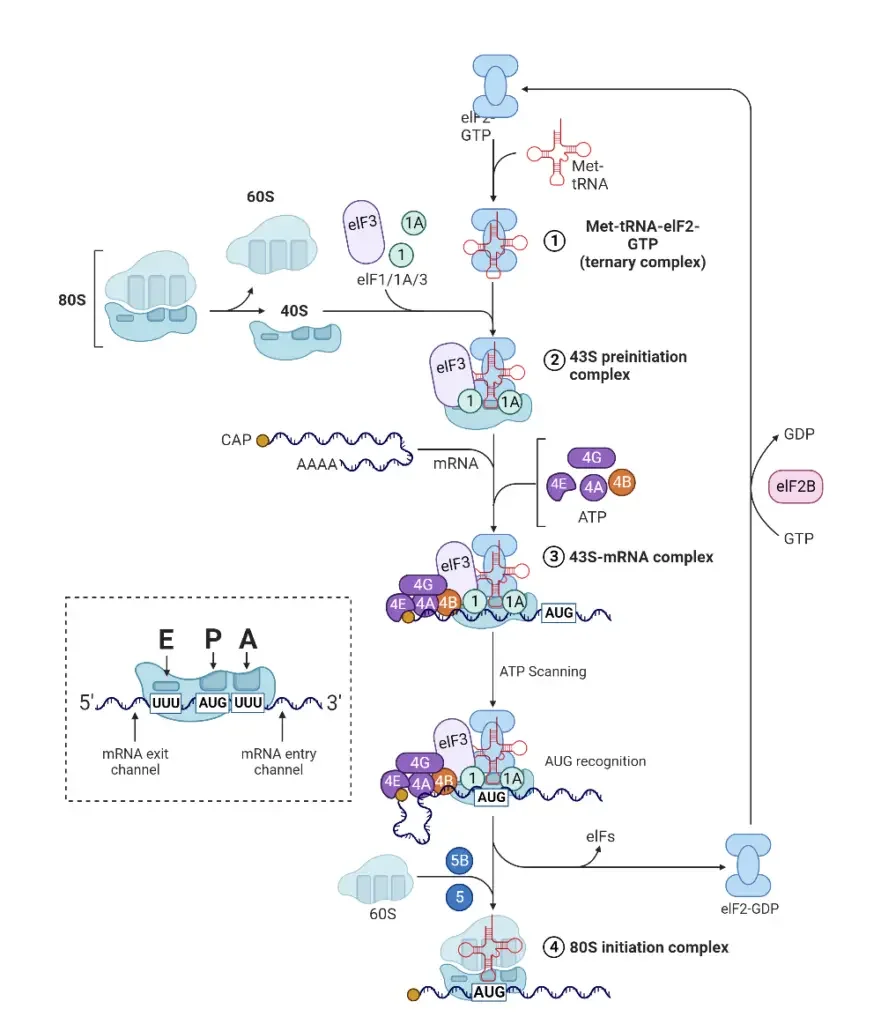
A. Translation Initiation
Translation initiation is the foundational step in protein synthesis, setting the stage for the accurate decoding of mRNA into a polypeptide chain. This process involves a series of coordinated interactions between various molecular components, ensuring the precise commencement of translation.
- Role of Initiation Factors The initiation of protein synthesis is orchestrated by several initiation factors, namely IF1, IF2, and IF3. These factors play pivotal roles in ensuring the correct assembly of the ribosome, mRNA, and tRNA, setting the stage for the subsequent elongation phase of translation.
- Binding of the Small Ribosomal Subunit The process begins with the small ribosomal subunit binding upstream on the 5′ end of the mRNA. Guided by the initiation factors, the ribosome scans the mRNA from the 5′ to 3′ direction. This scanning continues until the ribosome encounters a start codon, which can be AUG, GUG, or UUG.
- Recognition by Initiator fMet-tRNA Upon identifying a start codon, the initiator fMet-tRNA, carrying the amino acid methionine (Met), recognizes and binds to it. This tRNA molecule is unique as it carries N-formylmethionine, signifying that methionine is the inaugural amino acid in the forming polypeptide chain. Despite its distinct form, the initiator fMet-tRNA possesses a conventional methionine anticodon, ensuring the insertion of N-formylmethionine at the beginning of the protein.
- Steps in Translation Initiation The initiation process can be broadly categorized into three sequential steps:
- Binding of mRNA to the Small Ribosomal Subunit: The small ribosomal subunit, with the assistance of initiator factor IF3, binds to the mRNA. This factor plays a crucial role in dissociating the ribosomal subunits, ensuring they exist as separate entities.
- Binding of Initiator fMet-tRNA: The initiator factor IF2, in conjunction with Guanine-triphosphate (GTP), facilitates the binding of the initiator fMet-tRNA to the P-site of the ribosome. This positions the tRNA for the subsequent addition of amino acids.
- Assembly of Ribosomal Subunits: A specific ribosomal protein hydrolyzes the GTP bound to IF2. This action provides the energy required for the assembly of the two ribosomal subunits. Following this, both IF3 and IF2 are released, marking the end of the initiation phase.
B. Translation Elongation
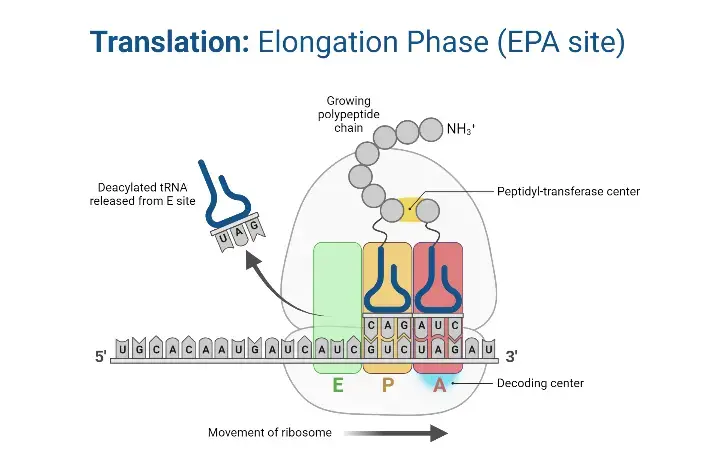
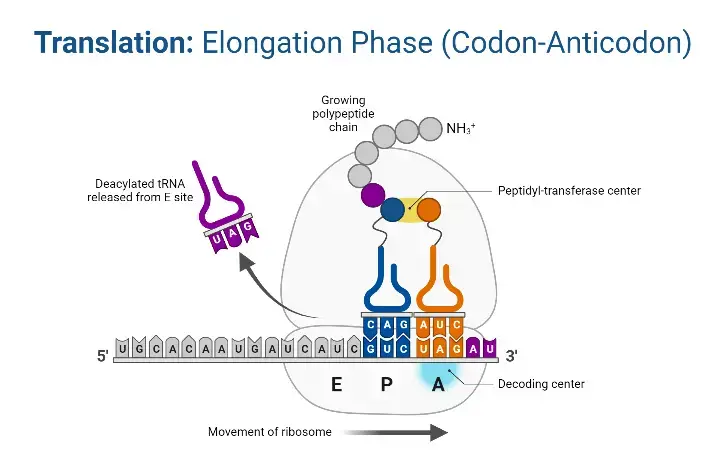
Translation elongation is the central phase of protein synthesis where the polypeptide chain is extended by the sequential addition of amino acids. This process is facilitated by the ribosome, which reads the mRNA codons and ensures the accurate placement of amino acids.
- Elongation Factors The elongation process is orchestrated by three primary protein factors: EF-Tu, EF-Ts, and EF-G. These factors play pivotal roles in ensuring the smooth and accurate addition of amino acids to the growing polypeptide chain.
- Ribosomal Function and Sites The ribosome operates by shifting one codon at a time, overseeing the activities within its three distinct sites: A (aminoacyl), P (peptidyl), and E (exit). During each step of elongation, a charged tRNA enters the ribosomal complex, contributing to the elongation of the polypeptide chain, while an uncharged tRNA exits. Remarkably, in prokaryotes, an amino acid is incorporated approximately every 0.05 seconds, translating to the synthesis of a 200 amino acid polypeptide in just 10 seconds.
- Energy Source The energy required for the formation of bonds between amino acids is derived from Guanosine Triphosphate (GTP), a molecule analogous to Adenosine Triphosphate (ATP).
- Steps in Translation Elongation The elongation process can be broadly categorized into three sequential steps:
- Aminoacyl-tRNA Entry: Mediated by elongation Factor-Tu (EF-Tu), aminoacyl-tRNAs are ushered into the A site of the ribosome. Initially, EF-Tu binds to GTP, forming an active EF-Tu-GTP complex that subsequently binds to tRNA. The hydrolysis of GTP to GDP releases energy, propelling the binding of aminoacyl-tRNA to the A site. Following this, EF-Tu departs from the complex.
- EF-Ts Mediation: Elongation factor EF-Ts facilitates the release of the EF-Tu-GDP complex from the ribosome and promotes the reformation of the active EF-Tu-GTP complex.
- Translocation and Peptide Bond Formation: During this phase, the polypeptide chain on the peptidyl-tRNA is transferred to the aminoacyl-tRNA situated in the A-site. This transfer reaction is catalyzed by peptidyl transferase. Subsequently, the ribosome advances one codon further on the mRNA, a process driven by elongation factor EF-G and powered by the hydrolysis of GTP to GDP. As the ribosome progresses, the uncharged tRNA is released from the P-site, and the newly formed peptidyl-tRNA transitions from the A-site to the P-site.
C. Translation Termination
Translation termination is the concluding phase of protein synthesis. This stage is marked by the cessation of the elongation process and the release of the newly synthesized polypeptide chain. The termination is precisely orchestrated to ensure the accurate completion of protein synthesis.
- Stop Codons The termination process is initiated when the ribosome encounters one of the three specific stop codons: UAA, UAG, or UGA. Unlike other codons, these stop codons do not code for amino acids. Instead, they signal the end of the translation process.
- Release Factors The stop codons are not recognized by tRNA molecules. Instead, they are identified by specialized protein factors known as release factors. Specifically:
- RF1: Recognizes and binds to the UAA and UAG stop codons.
- RF2: Recognizes and binds to the UAA and UGA stop codons.
- RF3: This is an auxiliary factor that aids in catalyzing the termination process, ensuring the efficient release of the polypeptide chain.
- Termination Process As the elongation phase progresses, the peptidyl-tRNA eventually reaches the P site of the ribosome. Upon encountering a stop codon, the corresponding release factor binds to the A site of the ribosome. This binding prompts the release of the polypeptide chain from the P site. Consequently, the ribosome dissociates into its two constituent subunits. This dissociation is an energy-dependent process, powered by the hydrolysis of GTP.
- Post-Termination Events After the termination of translation, the mRNA, having been read multiple times by successive ribosomes, undergoes degradation. This degradation process recycles the nucleotides, making them available for subsequent transcription reactions.
Explain the roles of mrna and trna in protein synthesis?
mRNA (messenger RNA) and tRNA (transfer RNA) are two crucial molecules involved in the process of protein synthesis, also known as translation. Protein synthesis occurs in the cell’s ribosomes and involves the conversion of the genetic information stored in DNA into functional proteins.
Here’s a breakdown of the roles of mRNA and tRNA in protein synthesis:
mRNA:
- Transcription: Initially, DNA is transcribed into mRNA in the cell nucleus. Enzymes called RNA polymerases create a complementary strand of mRNA using one of the DNA strands as a template. This process occurs during the first stage of gene expression, known as transcription.
- mRNA Structure: The mRNA molecule is composed of a series of nucleotides linked together. Each nucleotide consists of a nitrogenous base (adenine, guanine, cytosine, or uracil), a ribose sugar, and a phosphate group. The sequence of these nucleotides determines the genetic code carried by the mRNA.
- Template for Translation: mRNA serves as a template during translation. It carries the genetic information from the DNA to the ribosomes in the cytoplasm or endoplasmic reticulum, where protein synthesis occurs.
- Codons: The sequence of nucleotides in mRNA is organized into three-letter codes called codons. Each codon specifies a particular amino acid or serves as a start or stop signal for protein synthesis. There are 64 possible codons, representing 20 different amino acids and three stop signals.
- Genetic Code: The genetic code is the set of rules that defines how codons are translated into specific amino acids. For example, the codon AUG codes for the amino acid methionine and also serves as the start codon for protein synthesis.
tRNA:
- Structure: tRNA is a relatively small RNA molecule that folds into a specific three-dimensional structure. It has an anticodon sequence on one end and an attachment site for amino acids on the other end.
- Anticodon: The anticodon is a three-nucleotide sequence on tRNA that is complementary to a specific codon on the mRNA. It allows tRNA to recognize and bind to the corresponding codon during translation. For example, if the codon on the mRNA is UAC, the anticodon on the tRNA would be AUG.
- Amino Acid Attachment: Each tRNA molecule is specific to a particular amino acid. At its attachment site, tRNA carries the corresponding amino acid that corresponds to the anticodon sequence. Enzymes called aminoacyl-tRNA synthetases are responsible for attaching the correct amino acid to its corresponding tRNA molecule.
- Delivery of Amino Acids: During translation, tRNA molecules bring their attached amino acids to the ribosome and align them according to the sequence of codons on the mRNA. The anticodon on the tRNA base-pairs with the corresponding codon on the mRNA, allowing the ribosome to join the amino acids together, forming a polypeptide chain.
Macromolecules involve in Protein synthesis
- Ribosomes:
- Eukaryotic ribosomes consist of two subunits: a 40S small subunit and a 60S large subunit, collectively forming an 80S ribosome.
- Ribosomes are ribonucleoprotein complexes comprising 4 RNAs and 80 proteins.
- They play a pivotal role in protein synthesis, with many functions attributed to ribosomal RNA (rRNA).
- Bacterial ribosomes are smaller, consisting of 30S and 50S subunits, forming a 70S particle.
- Eukaryotic cells also contain mitochondrial ribosomes, which have origins traceable to bacteria.
- Messenger RNA (mRNA):
- mRNA carries the coding section of a gene essential for protein synthesis.
- It contains both non-coding and coding sequences, with the coding sequence grouping nucleotides into codons.
- Transfer RNA (tRNA):
- tRNAs act as adaptors, connecting the nucleotide sequence in mRNAs to the amino acid sequence in proteins.
- They have a cloverleaf-like structure with an amino acid linked at the 3’ end and an anticodon at the base.
- Aminoacyl-tRNA synthetases charge tRNAs by linking them to their respective amino acids.
- Genetic Code:
- The genetic code comprises three nucleotides that specify the amino acids in proteins.
- It is degenerate, meaning multiple codons can specify a single amino acid.
- Sixty-one codons specify amino acids, while three serve as “stop” codons.
- Some deviations from the universal code exist in mitochondria and specific bacterial species.
- Genetic Code and Human Disease:
- Proper readout of the genetic code is vital for human health.
- Mutations can be synonymous (no change in amino acid) or nonsynonymous (change in amino acid).
- Nonsynonymous mutations include missense, nonsense, and frameshift mutations.
- Protein Factors:
- Protein synthesis involves multiple non-ribosomal proteins.
- These proteins function transiently during initiation, elongation, and termination phases.
- They are named based on their function, such as eukaryotic initiation factor 2 (eIF2).
Testing of protein synthesis
- Immunostaining:
- Purpose: Immunostaining is a technique used to detect the presence of specific proteins in a clinical specimen.
- Procedure: The specimen is treated with a primary antibody solution that targets the protein of interest. This antibody may carry a fluorescent molecule or an enzyme that produces fluorescence when exposed to a specific substrate.
- Visualization: The emitted light can be observed under a microscope or captured on photosensitive film in a dark room.
- Types: There are two main types of immunostaining:
- Direct Immunostaining: The primary antibody itself is fluorescently labeled.
- Indirect Immunostaining: A secondary antibody, which targets the primary antibody, is used for fluorescent detection.
- Protein Electrophoresis:
- Purpose: Protein electrophoresis is a method used to separate proteins based on their size and/or charge.
- Procedure: Proteins can be run in their natural state or can be denatured using detergents like sodium dodecyl sulfate (SDS). Denaturing breaks non-covalent bonds within proteins and provides them with a uniform charge-to-mass ratio.
- Separation Mechanism: In SDS-polyacrylamide gel electrophoresis, the only factor influencing protein movement is the molecular sieving action of the polyacrylamide gel.
- Detection: After separation, proteins can be identified in several ways:
- Non-specific Detection: Using dyes like coomassie blue which stain all proteins.
- Specific Detection: Using antibodies in a method known as Western blotting or immunoblotting.
Pathophysiology of protein synthesis
Protein synthesis is a vital cellular process that translates genetic information into functional proteins. However, mutations or defects in this process can lead to various human diseases, either due to altered protein sequences or defects in the protein synthetic machinery itself.
- Sickle Cell Anemia:
- Description: Sickle Cell Anemia is a genetic disorder affecting hemoglobin, the oxygen-carrying protein in red blood cells.
- Cause: A missense mutation in the sixth codon of the beta chain of hemoglobin results in the substitution of glutamic acid with valine.
- Effect: This mutation alters the tertiary and quaternary structures of hemoglobin, causing red blood cells to adopt a sickle shape under certain conditions, leading to various complications.
- Duchenne Muscular Dystrophy (DMD):
- Description: DMD is an X-linked disease primarily affecting males, characterized by muscle weakness and other clinical symptoms.
- Cause: A premature stop codon in an early exon of the dystrophin gene leads to the production of a truncated dystrophin protein.
- Effect: The compromised dystrophin protein affects the integrity of the sarcomere and the muscle’s contractile function.
- Diamond-Blackfan Anemia (DBA):
- Description: DBA is a condition resulting from defects in the protein synthetic machinery rather than mutations in protein-coding sequences.
- Cause: Autosomal dominant mutations in genes encoding proteins of either the 40S or 60S ribosomal subunit are responsible for DBA.
- Effect: The exact pathophysiological mechanisms of DBA remain unclear. However, it is believed that changes in the cellular proteome due to suboptimal ribosome numbers contribute to the disease’s clinical features, including red blood cell production deficit, small stature, and various congenital anomalies.
What is the first step of protein synthesis?
The first step of protein synthesis is transcription. It is the process by which genetic information encoded in DNA is copied into a complementary RNA molecule called messenger RNA (mRNA). Transcription occurs in the nucleus of eukaryotic cells and in the cytoplasm of prokaryotic cells.
Here is a summary of the steps involved in transcription:
- Initiation: Transcription begins with the binding of RNA polymerase enzyme to the DNA template strand at a specific region called the promoter. The promoter sequence helps to determine the start site for transcription. Transcription factors, proteins that regulate gene expression, assist in the binding of RNA polymerase to the promoter.
- Elongation: RNA polymerase unwinds the DNA double helix, exposing a small stretch of the template strand. It then adds nucleotides to the growing RNA molecule, complementary to the exposed DNA template strand. The RNA molecule is synthesized in the 5′ to 3′ direction, using ribonucleotide triphosphates (ATP, GTP, CTP, and UTP) as building blocks. As RNA polymerase moves along the DNA template, the newly synthesized RNA molecule detaches from the DNA template.
- Termination: Transcription continues until a termination signal is reached in the DNA template. This signal prompts RNA polymerase to detach from the DNA template strand, releasing the newly synthesized RNA molecule. In prokaryotes, termination signals may involve specific DNA sequences. In eukaryotes, additional factors are involved in the termination of transcription.
After transcription, the mRNA molecule undergoes additional processing steps in eukaryotes. These include the removal of non-coding regions called introns and the joining together of coding regions called exons through a process called RNA splicing. The processed mRNA molecule is then transported out of the nucleus into the cytoplasm, where it serves as a template for translation, the next step in protein synthesis.
In summary, the first step of protein synthesis is transcription, where the genetic information in DNA is transcribed into mRNA molecules.
What is the second step of protein synthesis?
The second step of protein synthesis is called translation. It follows the first step, which is transcription. During translation, the information encoded in the mRNA molecule is used to assemble a chain of amino acids, forming a polypeptide chain or protein. The process of translation involves several key components, including ribosomes, transfer RNA (tRNA), and amino acids.
Here is a summary of the steps involved in translation:
- Initiation: The small ribosomal subunit binds to the mRNA molecule near the start codon (usually AUG) with the help of initiation factors. The initiator tRNA, carrying the amino acid methionine (or formylmethionine in prokaryotes), binds to the start codon in the ribosome’s P site.
- Elongation: The ribosome moves along the mRNA molecule in a 5′ to 3′ direction. A new tRNA carrying the appropriate amino acid binds to the codon in the ribosome’s A site, complementary to the mRNA codon. The ribosome catalyzes the formation of a peptide bond between the amino acid in the P site and the amino acid in the A site, creating a growing polypeptide chain. The ribosome then translocates, shifting by one codon, with the tRNA in the P site moving to the E site (exit site) and the tRNA in the A site moving to the P site.
- Termination: When a stop codon (UAA, UAG, or UGA) is encountered in the mRNA, it signals the end of translation. Release factors bind to the stop codon, causing the release of the completed polypeptide chain from the ribosome. The ribosome subunits dissociate, and the mRNA and newly synthesized protein are released.
It’s important to note that while transcription occurs in the nucleus of eukaryotic cells, translation takes place in the cytoplasm, where ribosomes are located. In prokaryotic cells, which lack a nucleus, transcription and translation can occur simultaneously in the cytoplasm.
What are the factors that affect protein synthesis?
Several factors can influence protein synthesis in cells. Here are some key factors that can affect the process:
- Availability of mRNA: The abundance and stability of mRNA molecules play a crucial role in protein synthesis. If there is a limited supply of mRNA encoding a particular protein, the synthesis of that protein may be reduced.
- Transcription factors: Transcription factors are proteins that bind to specific DNA sequences and regulate the transcription of genes. The activity of transcription factors can influence the rate of mRNA synthesis, thereby affecting protein synthesis.
- Availability of amino acids: Adequate availability of amino acids is essential for protein synthesis. Cells need a sufficient supply of all the necessary amino acids to synthesize proteins accurately and efficiently.
- Energy supply: Protein synthesis is an energy-intensive process that requires ATP (adenosine triphosphate). If cellular energy levels are low or ATP production is compromised, protein synthesis may be hindered.
- Post-transcriptional modifications: Various post-transcriptional modifications, such as alternative splicing and RNA editing, can affect mRNA stability and translation efficiency. These modifications can influence which parts of the mRNA are translated and ultimately impact protein synthesis.
- Ribosome availability: Ribosomes are the cellular machinery responsible for protein synthesis. The number and availability of ribosomes can impact the overall rate of protein synthesis. Factors that affect ribosome biogenesis, such as nutrient availability and cellular stress, can influence protein synthesis.
- Regulatory molecules: Certain molecules, such as microRNAs (miRNAs) and other non-coding RNAs, can regulate protein synthesis by binding to mRNA molecules and inhibiting their translation. Additionally, proteins called translational regulators can affect the efficiency and rate of protein synthesis by interacting with ribosomes or mRNA.
- Environmental conditions: External factors, such as temperature, pH, and the presence of stressors (e.g., toxins, heat shock, nutrient deprivation), can affect protein synthesis. Cells may respond to such conditions by altering gene expression, including the regulation of protein synthesis.
- Hormones and signaling pathways: Hormones and signaling molecules can modulate protein synthesis by activating or repressing specific signaling pathways. These pathways can regulate the expression of genes involved in protein synthesis, influencing the overall process.
- Cell cycle and developmental stage: Protein synthesis can be tightly regulated during different stages of the cell cycle and during development. The rates and types of proteins synthesized may vary depending on the specific cell cycle phase or developmental context.
Protein Synthesis Inhibitors
Protein synthesis inhibitors are compounds that disrupt the process of protein synthesis, which is vital for cell growth and replication. These inhibitors target specific stages or components of the protein synthesis machinery, leading to the cessation or alteration of protein production.
- Antibiotics as Inhibitors
- Puromycin: An antibiotic that mimics the structure of aminoacyl-tRNA. It prematurely terminates protein synthesis by incorporating itself into the growing polypeptide chain, leading to its release from the ribosome.
- Streptomycin: A trisaccharide antibiotic that affects the binding of formyl methionyl-tRNA to ribosomes, preventing the correct initiation of protein synthesis.
- Aminoglycosides: This group, including neomycin, kanamycin, and gentamycin, interferes with the decoding site in the 16S rRNA of the small ribosomal subunit.
- Chloramphenicol: Specifically inhibits the peptidyl transferase activity, halting the elongation of the polypeptide chain.
- Erythromycin: A macrolide antibiotic that blocks the translocation step by binding to the 50S ribosomal subunit.
- Tetracyclines: Bind to the 30S ribosomal subunit, preventing the binding of aminoacyl-tRNA, and thus inhibiting protein elongation.
- Toxins as Inhibitors
- Diphtheria toxin: This toxin’s A fragment catalyzes the transfer of a single side chain of elongation factor EF2, blocking the translocation of the growing polypeptide chain.
- Ricin: A potent toxin that inactivates the 60S ribosomal subunit by removing an adenine residue from rRNA, leading to premature termination of protein synthesis.
- Chemotherapeutic Agents
- Cycloheximide: Commonly used in laboratory settings, it blocks protein synthesis in eukaryotic cells by inhibiting the peptidyl transferase activity of ribosomes.
- Other Inhibitors
- mTOR Inhibitors: Such as Rapamycin, these inhibit the protein kinase mTOR, leading to reduced protein synthesis.
- Proteasome Inhibitors: Bortezomib, for instance, inhibits the 26S proteasome involved in protein degradation. The accumulation of abnormal proteins can lead to cell death.
| Type | Inhibitor | Description |
|---|---|---|
| Antibiotics | Puromycin | Mimics aminoacyl-tRNA structure, leading to premature termination of protein synthesis. |
| Streptomycin | Affects the binding of formyl methionyl-tRNA to ribosomes, preventing correct initiation. | |
| Aminoglycosides (e.g., neomycin, kanamycin, gentamycin) | Interferes with the decoding site in the 16S rRNA of the small ribosomal subunit. | |
| Chloramphenicol | Inhibits the peptidyl transferase activity. | |
| Erythromycin | Blocks the translocation step by binding to the 50S ribosomal subunit. | |
| Tetracyclines | Bind to the 30S ribosomal subunit, preventing the binding of aminoacyl-tRNA. | |
| Toxins | Diphtheria toxin | Inactivates elongation factor EF2, halting protein elongation. |
| Ricin | Inactivates the 60S ribosomal subunit by removing an adenine residue from rRNA. | |
| Chemotherapeutic Agents | Cycloheximide | Inhibits the peptidyl transferase activity of ribosomes. |
| mTOR Inhibitors (e.g., Rapamycin) | Inhibits the protein kinase mTOR, leading to reduced protein synthesis. | |
| Other Inhibitors | Proteasome Inhibitors (e.g., Bortezomib) | Inhibits the 26S proteasome involved in protein degradation. |
Eukaryotes Protein Synthesis vs. Prokaryotes Protein Synthesis
| Features | Eukaryotes | Prokaryotes |
|---|---|---|
| mRNA Type | Monocistronic, coding for a single gene of polypeptides | Polycistronic, coding for several genes of polypeptides |
| RNA Polymerase | Three types | Single type |
| Ribosomes | 40S and 60S subunits | 70S ribosomes |
| Transcription/Translation Overlap | Separate processes | Processes can overlap |
| mRNA Modification | Pre-mRNA undergoes modifications (splicing, 5′ cap, poly-A tail) | Limited modification |
| Initiator tRNA | Special initiator complex | Special initiator tRNA (Met-tRNAf or Met-tRNA) |
| Starting Amino Acid | Methionine | N-formyl methionine |
| Initiation/Termination Sites | Single site | Multiple sites |
| Ribosomal Binding Site | Kozak sequence centered around start codon | Shine-Dalgarno sequence (-10 nucleotides upstream of initiation codon) |
| Initiation Factors | Several (eIF-2, eIF-2al, eIF-a2, eIF-a) | IF-1, IF-2, IF-3 |
| Chain Elongation Factors | EF-1 and EF-2 | EF-Tu, EF-Ts, EF-G |
| Release Factors | Single (eRF) for recognition of all stop codons (UAA, UAG, UGA) | Multiple (RF-1, RF-2, RF-3) for recognition of stop codons |
| Mitochondrial/Chloroplast Genetic Code | Can differ within eukaryotes | Same in all prokaryotes |
FAQ
_ are the sites of protein synthesis.
The cytoplasm and the rough endoplasmic reticulum (ER) are the sites of protein synthesis.
the process of protein synthesis is called __.
The process of protein synthesis is called translation.
What is protein synthesis?
Protein synthesis refers to the biological process by which cells generate proteins. It involves the conversion of genetic information stored in DNA into functional proteins.
Where does protein synthesis occur in the cell?
Protein synthesis takes place in the ribosomes, which can be found in the cytoplasm of the cell or attached to the endoplasmic reticulum (ER) in eukaryotic cells.
What is the first step of protein synthesis?
The first step of protein synthesis is transcription, during which DNA is transcribed into mRNA. This process occurs in the cell nucleus.
What is the role of mRNA in protein synthesis?
mRNA carries the genetic information from DNA to the ribosomes. It serves as a template for translation and carries the codons that specify the sequence of amino acids in a protein.
What is the genetic code in protein synthesis?
The genetic code is a set of rules that determines how codons (three-letter sequences of nucleotides) in mRNA are translated into specific amino acids during protein synthesis.
What is the role of tRNA in protein synthesis?
tRNA molecules bring the appropriate amino acids to the ribosomes during translation. They recognize and bind to the codons on mRNA through their anticodon sequence and deliver the corresponding amino acids to build the protein chain.
How are amino acids joined together during protein synthesis?
Amino acids are joined together by peptide bonds. Ribosomes catalyze the formation of these bonds between adjacent amino acids to build the protein chain.
What is the role of ribosomes in protein synthesis?
Ribosomes serve as the site of protein synthesis. They provide the platform where mRNA and tRNA interact, enabling the assembly of amino acids into a polypeptide chain.
What is the stop codon in protein synthesis?
Stop codons are specific codons (UAA, UAG, and UGA) on mRNA that signal the termination of protein synthesis. When a ribosome encounters a stop codon, it releases the completed protein chain.
What happens after protein synthesis is complete?
After protein synthesis, the newly synthesized protein undergoes various post-translational modifications, such as folding, modifications of amino acid residues, and targeting to specific cellular compartments. These processes are essential for the protein to acquire its functional structure and perform its specific role in the cell.
References
- Hoerter JE, Ellis SR. Biochemistry, Protein Synthesis. [Updated 2023 Jul 17]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK545161/
- https://www.biologyonline.com/dictionary/protein-synthesis
- https://humanbiology.pressbooks.tru.ca/chapter/5-6-protein-synthesis/
- https://biologydictionary.net/protein-synthesis/