What is Prokaryotic Translation?
In the realm of molecular biology and genetics, the synthesis of proteins is a pivotal process known as translation. This process is executed by ribosomes, either in the cytoplasm or the endoplasmic reticulum. It is essential to note that translation follows the transcription of DNA to RNA within the cell’s nucleus, collectively termed gene expression.
Therefore, during the intricate process of translation, the ribosome decodes the messenger RNA (mRNA) to produce a specific chain of amino acids, referred to as a polypeptide. This polypeptide subsequently undergoes folding to transform into an active protein, which then carries out its designated functions within the cell. Besides the formation of the polypeptide, translation is responsible for transferring the information encapsulated in mRNA molecules into the sequences of amino acids in the resultant gene products.
While translation is a universal cellular mechanism, it is imperative to understand that its process varies between prokaryotic and eukaryotic cells. Then, for the purpose of this exposition, the focus will be on prokaryotic translation.
Prokaryotic translation predominantly takes place within the cytosol. Here, the large and small subunits of the ribosome attach themselves to the mRNA, initiating the translation process. This process can be further divided into three sequential phases: Initiation, Elongation, and Termination.
- Initiation Phase: During this initial phase, the ribosome meticulously assembles around the target mRNA. The first transfer RNA (tRNA) is strategically positioned at the start codon, marking the beginning of the translation.
- Elongation Phase: As the process progresses to the elongation phase, the preceding tRNA, after validation by the small ribosomal subunit, transfers its carried amino acid to the large ribosomal subunit. This large subunit then binds the amino acid to one of the previously admitted tRNA through a process called transpeptidation. Subsequently, the ribosome advances to the next mRNA codon through translocation, facilitating the formation of the amino acid chain.
- Termination Phase: The culmination of the translation process is marked by the termination phase. Here, upon encountering a stop codon, the ribosome releases the synthesized polypeptide. However, the ribosomal complex remains intact, poised to commence the translation of the next mRNA.
Component Required for Prokaryotic Translation
- Amino Acids: Proteins are essentially polymers composed of amino acids. Out of the 20 amino acids integral to protein structure, humans can synthesize only half. The remaining 10, termed essential amino acids, must be sourced from the diet. For successful protein synthesis, the presence of all requisite amino acids is mandatory. If even one essential amino acid is deficient in the diet, translation halts. Therefore, a consistent dietary supply of these amino acids is crucial. However, prokaryotes, unlike humans, can synthesize all 20 amino acids from inorganic components.
- Ribosomes: Often referred to as the factories of protein synthesis, ribosomes are pivotal to translation. Comprising roughly equal parts of protein and RNA, ribosomes are intricate structures. In prokaryotes, they are denoted as 70S, with two subunits: a larger one and a smaller one. These subunits dissociate post-translation and reassociate during initiation. The ribosome has two primary sites, the A site (aminoacyl tRNA binding) and the P site (peptidyl tRNA binding). Additionally, ribosomes are associated with the rough endoplasmic reticulum, forming clusters where protein synthesis transpires.
- Messenger RNA (mRNA): mRNA carries the specific information necessary for synthesizing a particular protein. It acts as a template, providing the sequence of amino acids for the polypeptide gene products.
- Transfer RNAs (tRNAs): Acting as the bridge between amino acids and the growing peptide chain, tRNAs transport amino acids. They possess an anticodon, a three-nucleotide base sequence, which recognizes the mRNA’s codon during protein synthesis. The diversity in tRNAs is vast, with humans having approximately 50 different types and bacteria around 40.
- Energy Sources: The energy-intensive process of translation requires both ATP and GTP. These molecules provide the necessary energy, with some reactions breaking down ATP or GTP to release pyrophosphate.
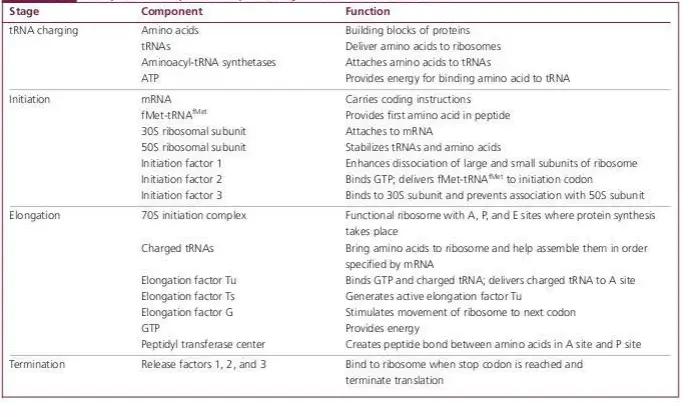
- Protein Factors: Translation is facilitated by various protein factors, each aiding in specific stages like initiation, elongation, and termination. These factors, listed in the table below, have distinct functions:
| Factor | Translation Step | Function |
|---|---|---|
| IF-1 | Initiation | Stabilizes 30S ribosomal subunit |
| IF-2 | Initiation | Binds fmet-tRNA with 30S subunit mRNA complex; binds and hydrolyzes GTP |
| IF-3 | Initiation | Binds 30S subunit with mRNA |
| EF-TU | Elongation | Binds GTP; brings Aminoacyl-tRNA to A-site of ribosome |
| EF-TS | Elongation | Generates EF-TU |
| EF-G | Elongation | Aids in ribosome translocation |
| RF-1 | Termination | Dissociates polypeptide from tRNA ribosome complex; specific for UAA and UAG |
| RF-2 | Termination | Dissociates polypeptide; specific for UGA and UAA |
| RF-3 | Termination | Stimulates RF-1 and RF-2 |
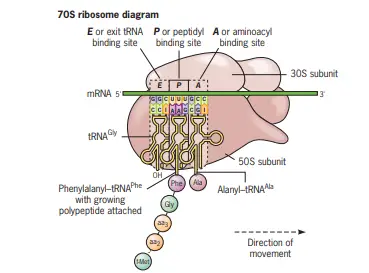
Ribosomal Sites for Protein Translation
Ribosomes play a pivotal role in protein synthesis, a process integral to cellular function. Within the prokaryotic ribosome, there are specific binding sites for transfer RNAs (tRNAs), each serving a unique purpose during translation. These sites are crucial for the systematic and sequential addition of amino acids to the growing polypeptide chain.
- Aminoacyl-tRNA Binding Site (A Site):
- Description: The A site, also known as the aminoacyl-tRNA binding site, is one of the three primary tRNA binding sites on the prokaryotic ribosome.
- Function: During the elongation phase of translation, the incoming aminoacyl-tRNA, carrying a specific amino acid, binds to this site. This ensures that the correct amino acid is added to the growing polypeptide chain based on the codon sequence of the messenger RNA (mRNA).
- Peptidyl-tRNA Binding Site (P Site):
- Description: The P site, or the peptidyl-tRNA binding site, is the second primary tRNA binding site on the ribosome.
- Function: This site holds the tRNA molecule that is linked to the growing polypeptide chain. As the chain elongates, the tRNA at the A site transfers its amino acid to the chain at the P site, ensuring the correct sequence of amino acids in the protein.
- Exit Site (E Site):
- Description: The E site, known as the exit site, is the third and final primary tRNA binding site on the ribosome.
- Function: After a tRNA molecule has played its role in translation by donating its amino acid to the growing chain, it moves to the E site. From here, the tRNA is subsequently released from the ribosome, making it available to be recharged with another amino acid and participate in subsequent rounds of translation.
- Role of rRNA in Forming Sites: Besides the protein components of the ribosome, ribosomal RNA (rRNA) molecules play a significant role in forming these tRNA binding sites. All three sites – A, P, and E – are structured by the rRNA molecules present in the ribosome. This emphasizes the importance of rRNA in ensuring the accuracy and efficiency of the translation process.
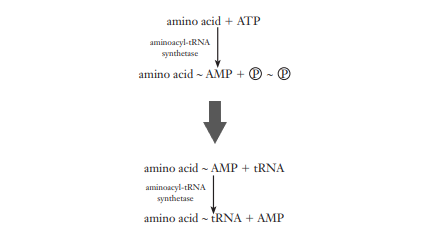
Activation of Amino acid/Synthesis of aminoacyl-tRNA
The activation of amino acids is a fundamental precursor to protein synthesis. This process occurs within the cytosol and is essential for the subsequent attachment of amino acids to their corresponding transfer RNAs (tRNAs). The activation and synthesis of aminoacyl-tRNA are two-step reactions that are vital for the translation process.
- Role of Aminoacyl-tRNA Synthetases: Aminoacyl-tRNA synthetases are a group of enzymes that play a critical role in the activation of amino acids. These enzymes exhibit high specificity for both the amino acid and the corresponding tRNA, ensuring the fidelity of protein synthesis. Each amino acid has a specific synthetase that catalyzes its attachment to the correct tRNA.
- Formation of Enzyme-AMP-Amino Acid Complex: The initial step in this process involves the amino acid being attached to the enzyme with the expenditure of ATP, forming an enzyme-AMP-amino acid complex. This reaction is the first stage of activation, where the amino acid is primed for transfer to the tRNA.
- Transfer to tRNA: Following the formation of the enzyme-AMP-amino acid complex, the amino acid is then transferred to the 3’ end of the tRNA, forming aminoacyl-tRNA. This step concludes the activation process, with the amino acid now linked to the tRNA by a high-energy bond, symbolized by a tilde (~).
- Function of Aminoacyl~tRNAs: The aminoacyl~tRNAs serve as substrates for polypeptide synthesis on ribosomes. They are responsible for recognizing the correct mRNA codon and presenting the amino acid in a three-dimensional structure conducive to peptide bond formation.
- Genetic Code Establishment and Activation: The linkage of an amino acid to a tRNA is crucial for establishing the genetic code. This attachment dictates the incorporation of the amino acid into the polypeptide chain based on the anticodon of the tRNA. Additionally, because peptide bond formation between free amino acids is not thermodynamically favorable, the amino acid must be activated, which is achieved through the formation of an amino acid ester with tRNA.
- Catalysis by Aminoacyl-tRNA Synthetases: The activation reaction is catalyzed by aminoacyl-tRNA synthetases, which facilitate the formation of an aminoacyl adenylate from an amino acid and ATP. This reaction results in aminoacyl-AMP, a mixed anhydride intermediate.
- Transfer Reaction: The aminoacyl group from the aminoacyl-AMP is then transferred to a specific tRNA molecule, forming aminoacyl-tRNA. This transfer is coupled with the release of AMP.
- Overall Reaction Energetics: The overall reaction, which includes the activation and transfer steps, is driven forward by the hydrolysis of pyrophosphate. This reaction is highly exergonic, consuming the equivalent of two ATP molecules for each aminoacyl-tRNA synthesized.
- Non-Dissociation of Aminoacyl-AMP: The aminoacyl-AMP intermediate remains tightly bound to the active site of the aminoacyl-tRNA synthetase through noncovalent interactions, preventing its dissociation during the synthesis of aminoacyl-tRNA.
- Importance of Aminoacyl-tRNA Synthesis: The synthesis of aminoacyl-tRNAs is critical for two main reasons: it allows amino acids to participate in protein synthesis by linking them to tRNA molecules, and it forms a high-energy bond that enables the amino acid to react with the growing polypeptide chain.
- tRNA Structure and Synthetase Specificity: Each tRNA molecule has a cloverleaf secondary structure with an accessible anticodon at the end of the anticodon stem loop. The amino acid is covalently bound to the A residue of the CCA sequence at the 3’ end of the tRNA. There are 20 different aminoacyl-tRNA synthetases, one for each amino acid, ensuring the specificity of the reaction.
Steps of Prokaryotic Translation
The Prokaryotic Translation is completed in three different steps such as;
- Polypeptide chain initiation.
- Chain elongation.
- Chain termination.
Initiation of Protein Synthesis
- Start Codon Recognition The initiation of protein synthesis in all mRNAs begins with the recognition of the start codon, which is universally the AUG codon. This codon encodes for the amino acid methionine, marking the starting point of polypeptide synthesis.
- tRNA Involvement Two distinct types of transfer RNA (tRNA) molecules are involved in interpreting the AUG codon. The initiator tRNA, known as tRNAfMet, is specifically used for the initiation codon. In contrast, tRNAm Met is utilized for AUG codons that occur internally within the mRNA sequence.
- First Amino Acid Incorporation in Prokaryotes In prokaryotic organisms, the first amino acid incorporated into a new protein is N-formylmethionine (fMet). Therefore, the aminoacyl-tRNA that is used during the initiation phase is fMet-tRNAfMet, which carries the modified methionine.
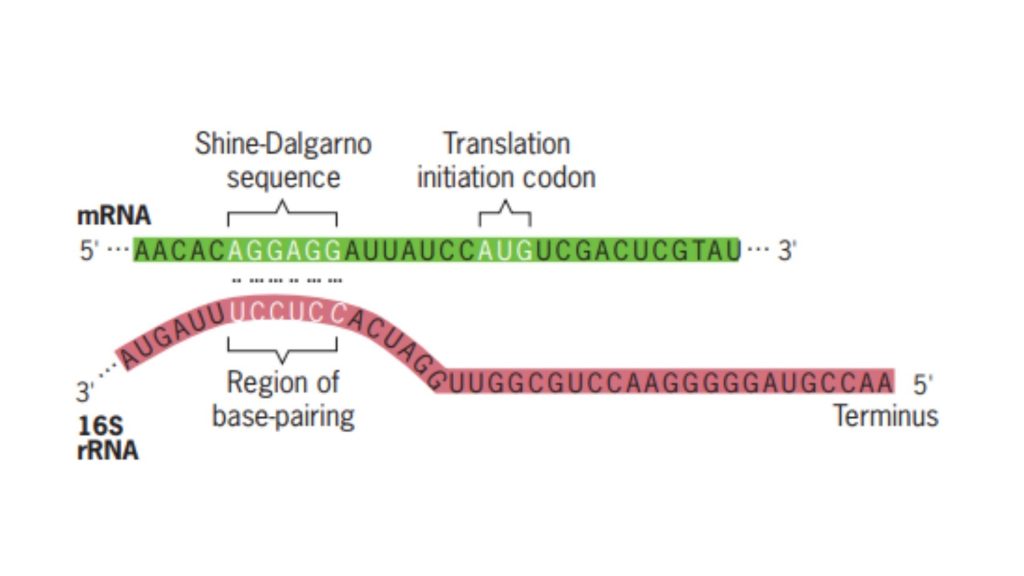
- Shine–Dalgarno Sequence A purine-rich sequence, designated as the Shine–Dalgarno sequence, is situated upstream of the AUG initiation codon. This sequence, 5’-AGGAGGU-3’, is complementary to a portion of the 16S rRNA in the small ribosomal subunit, thus serving as the binding site for the 30S ribosomal subunit. The 30S subunit binds here and then migrates in the 3’ direction along the mRNA until it locates the AUG initiation codon.
- Initiation Factors (IFs) Requirement The initiation of protein synthesis necessitates the involvement of proteins known as initiation factors (IFs). In prokaryotic systems, three initiation factors—IF-1, IF-2, and IF-3—are essential for the successful initiation of protein synthesis.
- Complexity of Initiation Factor Binding Due to the complexity of the initiation process, the precise order in which IF-1, IF-2, IF-3, and fMet-tRNAfMet bind to the ribosomal subunit is a subject of scientific inquiry and has not been definitively established. Each factor plays a critical role in ensuring the correct assembly of the initiation complex and the accurate start of protein synthesis.
Steps Involved in Initiation of Protein Synthesis
- Binding of Initiation Factors to the Small Ribosomal Subunit The initiation process commences with the binding of initiation factors IF-1 and IF-3 to the small ribosomal subunit, specifically the 30S subunit. Their primary function is to ensure the accurate assembly of the ribosomal subunits.
- Prevention of Premature Ribosomal Subunit Association IF-1 and IF-3 play a crucial role in preventing the 30S subunit from binding to the 50S subunit in the absence of mRNA and fMet-tRNAf Met. Such a premature association would lead to the formation of a nonfunctional ribosome.
- Binding of the Small Subunit to mRNA The small ribosomal subunit, after associating with the initiation factors, binds to the mRNA. This binding is facilitated by the Shine–Dalgarno sequence, a purine-rich sequence located upstream of the AUG initiation codon. The 30S subunit then moves in the 3’ direction along the mRNA until it identifies the AUG initiation codon.
- Initiator tRNA Binding The initiator tRNA, which is charged with the amino acid N-formylmethionine, forms a complex with initiation factor IF-2 and guanosine triphosphate (GTP). This complex, denoted as fMet-tRNAfMet/IF-2/GTP, subsequently binds to the initiation site on the ribosome.
- Release of IF-3 Upon the binding of the initiator tRNA complex, IF-3 is released from the ribosomal subunit. This step ensures that the ribosome is ready for the next phase of the initiation process.
- Formation of the 30S Initiation Complex The assembly comprising the mRNA, fMet-tRNAf Met, IF-1, IF-2, and the 30S ribosomal subunit is termed the 30S initiation complex. This complex is a precursor to the fully functional ribosome that will carry out protein synthesis.
- Binding of the Large Ribosomal Subunit The final step in the initiation phase involves the binding of the large ribosomal subunit, the 50S subunit. This binding event is accompanied by the release of initiation factors IF-1 and IF-2 and the hydrolysis of GTP. The resulting complex, known as the 70S initiation complex, signifies the completion of the initiation phase and the readiness of the ribosome to commence the elongation phase of protein synthesis.
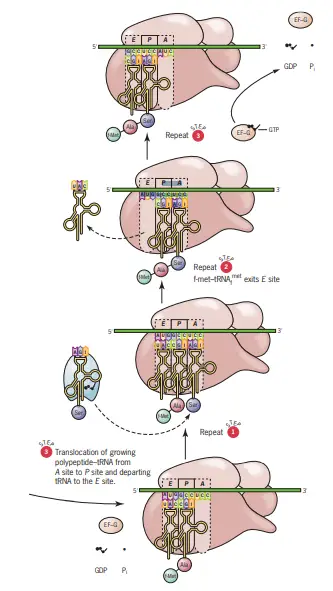
Elongation of Protein Synthesis
- Initial Positioning of the Initiation Codon At the onset of elongation, the initiation codon, typically AUG, is strategically positioned in the P site. Bound to this codon via codon–anticodon base pairing is the fMet-tRNAfMet. Subsequently, the mRNA’s next codon aligns itself in the A site, setting the stage for the elongation cycle.
- Elongation Cycle Overview The elongation of the polypeptide chain is a meticulous process that encompasses three primary steps: aminoacyl-tRNA binding, peptide bond formation, and translocation.
- Aminoacyl-tRNA Binding
- The specific aminoacyl-tRNA corresponding to the second codon in the mRNA sequence binds to the A site through codon–anticodon interactions.
- This binding process necessitates the involvement of the elongation factor EF-Tu and guanosine triphosphate (GTP), which together form an aminoacyl-tRNA/EF-Tu/GTP complex.
- Post binding, GTP undergoes hydrolysis, leading to the release of EF-Tu, now associated with GDP.
- To participate in subsequent rounds of elongation, EF-Tu must be regenerated. This regeneration, termed the EF-Tu–EF-Ts exchange cycle, involves the elongation factor EF-Ts. Initially, EF-Ts binds to EF-Tu, displacing the GDP. Following this, GTP binds to EF-Tu, ousting EF-Ts. The regenerated EF-Tu-GTP is then primed for another elongation cycle.
- Peptide Bond Formation
- The enzymatic action of peptidyl transferase facilitates the second step, peptide bond formation.
- During this reaction, the carboxyl end of the amino acid tethered to the tRNA in the P site detaches from the tRNA. It then forms a peptide bond with the amino group of the amino acid linked to the tRNA situated in the A site.
- Translocation
- The third step involves the binding of a complex comprising the elongation factor EF-G (also known as translocase) and GTP to the ribosome.
- Three synchronized movements, collectively termed translocation, ensue:
- The deacylated tRNA transitions from the P site to the E site.
- The dipeptidyl-tRNA in the A site relocates to the P site.
- The ribosome advances along the mRNA in a 5’ to 3’ direction by three nucleotides, positioning the subsequent codon in the A site.
- During these translocation events, GTP is hydrolyzed to GDP and inorganic phosphate, resulting in the release of EF-G. This prepares EF-G to bind more GTP for subsequent elongation cycles.
- Post-translocation, the A site remains vacant, primed to accommodate the next aminoacyl-tRNA. It’s noteworthy that the A and E sites cannot be simultaneously occupied. Hence, before the next aminoacyl-tRNA binds to the A site, the deacylated tRNA departs from the E site.
- Continuation of Elongation Elongation persists, appending one amino acid to the C-terminal end of the burgeoning polypeptide for each codon interpreted. The peptidyl-tRNA oscillates between the P and A sites as the polypeptide chain expands.
Detailed Steps of Elongation in Protein Synthesis
The elongation of the polypeptide chain is a fundamental process in protein synthesis, consistent across both prokaryotes and eukaryotes. This process involves the sequential addition of amino acids to the growing polypeptide chain, facilitated by a series of intricate steps.
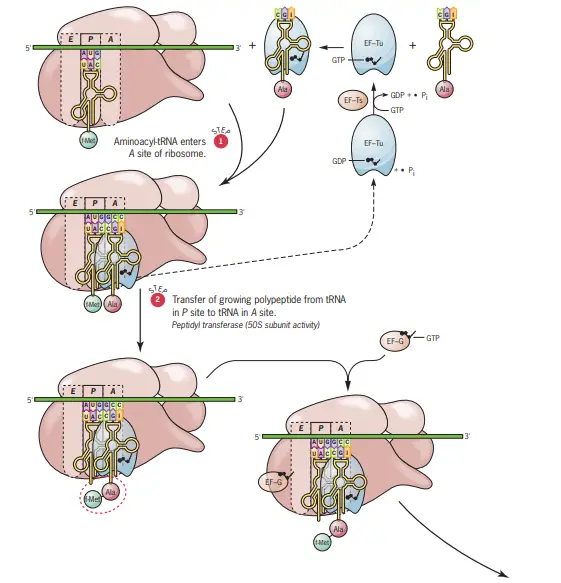
- Step One: Aminoacyl-tRNA Binding to the A Site
- The initial step involves the binding of an aminoacyl-tRNA to the A site of the ribosome.
- The specificity of this binding is determined by the mRNA codon aligned with the A site.
- The anticodon of the incoming aminoacyl-tRNA must form complementary base pairs with the mRNA codon at the A site.
- This binding process necessitates the involvement of the elongation factor Tu, associated with a GTP molecule (EF-Tu.GTP).
- While GTP is essential for the binding of aminoacyl-tRNA at the A site, its hydrolysis occurs only after the formation of the peptide bond.
- Post GTP hydrolysis, the ribosome releases EF-Tu now bound to GDP.
- The inactive EF-Tu.GDP is reactivated to EF-Tu.GTP by the elongation factor Ts (EF-Ts), which utilizes a GTP molecule in the process.
- Notably, EF-Tu interacts with all aminoacyl-tRNAs except for methionyl-tRNA.
- Step Two: Peptide Bond Formation
- The subsequent step involves the formation of a peptide bond. This bond is established between the amino group of the aminoacyl-tRNA in the A site and the carboxyl terminus of the growing polypeptide chain attached to the tRNA in the P site.
- This reaction results in the detachment of the growing chain from the tRNA in the P site and its attachment to the tRNA in the A site.
- The peptide bond formation is catalyzed by peptidyl transferase, an intrinsic enzymatic activity of the 50S ribosomal subunit.
- Intriguingly, the peptidyl transferase activity is attributed to the 23S rRNA molecule and not a ribosomal protein, hinting at the evolutionary significance of RNA in early life forms.
- The energy for this reaction is derived from the hydrolysis of the GTP molecule introduced by EF-Tu in the first step.
- Step Three: Ribosomal Translocation
- The third and final step of the elongation cycle involves the translocation of the ribosome along the mRNA.
- During this phase, the peptidyl-tRNA in the A site is shifted to the P site, while the uncharged tRNA in the P site moves to the E site. Concurrently, the ribosome advances by three nucleotides towards the 3′ end of the mRNA.
- This translocation requires the presence of GTP and the elongation factor G (EF-G).
- The ribosome undergoes conformational changes during translocation, suggesting a possible shuttling mechanism along the mRNA.
- The energy driving the ribosomal movement is sourced from the hydrolysis of GTP.
- Upon the completion of translocation, the A site becomes vacant, positioning the ribosome to initiate the next elongation cycle.
Termination of Protein Synthesis
The termination phase of protein synthesis is initiated when one of the three termination codons, also known as Stop codons, aligns with the A site of the ribosome. The three recognized termination codons are UAG, UAA, and UGA.
- Absence of Corresponding Aminoacyl-tRNAs Distinctively, these termination codons do not have corresponding aminoacyl-tRNAs in prokaryotic cells. This means that when these codons are encountered, the usual tRNA-mediated addition of amino acids to the growing polypeptide chain ceases.
- Role of Release Factors In the absence of aminoacyl-tRNAs for the Stop codons, specialized proteins called release factors come into play:
- RF-1 and RF-2: These are the primary release factors that recognize and bind to the termination codons. Specifically, RF-1 identifies and binds to UAA and UAG, while RF-2 recognizes and binds to UAA and UGA.
- RF-3: This is an auxiliary release factor that aids in the interaction of RF-1 or RF-2 with the ribosome. Depending on the specific termination codon present in the A site, either the combination of RF-1 + RF-3 or RF-2 + RF-3 will bind to the ribosome. While RF-1 (or RF-2) typically binds at or near the A site, RF-3, in association with GTP, binds to a different location on the ribosome.
- Action of Release Factors and Peptidyl Transferase The binding of the release factors triggers the peptidyl transferase activity of the ribosome. However, instead of facilitating the addition of another amino acid, the release factors induce peptidyl transferase to transfer the growing polypeptide chain to a water molecule. This action effectively severs the bond between the polypeptide and the tRNA situated in the P site.
- Dissociation and Release Following the cleavage, the newly synthesized polypeptide is released from the ribosome. Subsequently, the mRNA and the uncharged tRNA also depart from the ribosome. The ribosome then disassembles into its constituent 30S and 50S subunits, rendering them available for another round of translation.
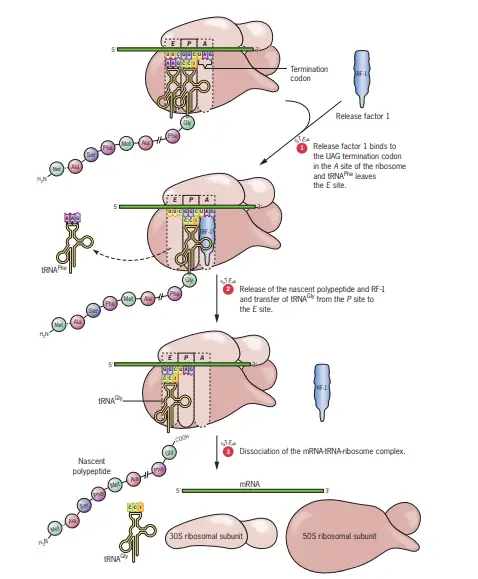
Steps of Termination of Protein Synthesis
- Encounter of Termination Codons: The ribosome reads the mRNA sequence and eventually encounters one of the three termination codons (Stop codons) in the A site. These codons are UAG, UAA, and UGA.
- Lack of tRNA Binding: Unlike regular codons, termination codons do not have corresponding aminoacyl-tRNAs in prokaryotic cells. As a result, no tRNA binds to these codons in the A site of the ribosome.
- Binding of Release Factors: In the absence of a corresponding tRNA, specialized proteins known as release factors recognize and bind to the termination codons:
- RF-1: Recognizes and binds to UAA and UAG.
- RF-2: Recognizes and binds to UAA and UGA. Depending on the specific termination codon present, either RF-1 or RF-2 will bind to the ribosome.
- Assistance of RF-3: An auxiliary release factor, RF-3, assists in the interaction of RF-1 or RF-2 with the ribosome. RF-3, in association with GTP, binds to a different location on the ribosome, ensuring the proper function of the primary release factors.
- Cleavage of the Polypeptide Chain: The binding of release factors triggers the ribosome’s peptidyl transferase activity. Instead of adding another amino acid, the release factors direct the peptidyl transferase to transfer the growing polypeptide chain to a water molecule. This action results in the cleavage of the bond between the polypeptide and the tRNA in the P site.
- Release of Components: After the polypeptide chain is cleaved and released, the mRNA and the uncharged tRNA also dissociate from the ribosome.
- Ribosome Dissociation: The ribosome subsequently disassembles into its two subunits, the 30S and 50S, making them available for another round of translation.
Components required for protein synthesis in bacterial cells
Post translation modification
After the synthesis of a polypeptide chain during translation, the protein often requires further modifications to become fully functional. This series of changes is referred to as post-translational modification.
- Amino Terminal and Carboxyl Terminal Modification:
- Bacteria: In bacterial proteins, the N-formylmethionine, which is the initial amino acid, is often removed from the polypeptide chain. Additionally, certain carboxyl terminal amino acids are enzymatically excised to produce a functional protein.
- Eukaryotes: In eukaryotic proteins, the amino terminal is typically N-acetylated, which is a common modification.
- Loss of Signal Sequences: Some proteins possess an amino terminal end known as a signal sequence. This sequence directs the protein to its proper cellular location. Once the protein reaches its destination, specific peptidases cleave off the signal sequence, rendering the protein without its signaling property.
- Modification of Individual Amino Acids: Individual amino acids within the protein can undergo various modifications to alter the protein’s function or stability. Common modifications include phosphorylation, where a phosphate group is added, and acetylation, where an acetyl group is added.
- Attachment of Carbohydrate Side Chains: Some proteins undergo glycosylation, where carbohydrate side chains are covalently attached. These proteins, known as glycoproteins, have diverse functions in the cell. Similarly, lipids can be attached to proteins, resulting in lipoproteins.
- Addition of Isoprenyl Group: Certain proteins require the addition of an isoprenyl group for activation. This lipid modification aids in anchoring the protein to cell membranes, enhancing its activity and interactions.
In conclusion, post-translational modifications are crucial for the proper functioning of proteins. These modifications ensure that proteins achieve the correct conformation, are directed to the appropriate cellular locations, and can interact effectively with other molecules. Therefore, understanding these modifications is essential for comprehending the diverse functions and regulations of proteins in the cell.
Difference Between Prokaryotic And Eukaryotic Translation
- Definition:
- Prokaryotic Translation: The processes of translation and transcription occur simultaneously in prokaryotes.
- Eukaryotic Translation: In eukaryotes, the translation and transcription processes are discontinuous, taking place in separate cellular compartments.
- Location of mRNA Synthesis:
- Prokaryotic: mRNA synthesis occurs in the cytoplasm.
- Eukaryotic: mRNA synthesis takes place inside the nucleus.
- Initiation Mechanism:
- Prokaryotic: Utilizes a cap-independent mechanism for translation initiation.
- Eukaryotic: Primarily uses a cap-dependent mechanism, although cap-independent mechanisms also exist.
- Ribosomal Structure:
- Prokaryotic: Translation is performed by 70S ribosomes, composed of 30S and 50S subunits.
- Eukaryotic: Translation is carried out by 80S ribosomes, made up of 40S and 60S subunits.
- mRNA Stability:
- Prokaryotic: mRNA molecules are generally unstable.
- Eukaryotic: mRNA molecules are more stable and have a longer lifespan.
- Lifespan of mRNA:
- Prokaryotic: The lifespan of mRNA ranges from a few seconds to 2 minutes.
- Eukaryotic: The mRNA can last from a few hours to several days.
- Occurrence in the Cell Cycle:
- Prokaryotic: Translation does not occur in a specific phase; it’s continuous.
- Eukaryotic: Translation primarily occurs during the G1 and G2 phases of the cell cycle.
- Rate of Process:
- Prokaryotic: The translation process is relatively fast.
- Eukaryotic: The translation process is slower in comparison.
- Release Factors:
- Prokaryotic: Utilizes release factors RF1 and RF2.
- Eukaryotic: Employs eukaryotic release factor, eRF.
- Initiation Factors:
- Prokaryotic: Uses three initiation factors.
- Eukaryotic: Utilizes nine distinct initiation factors.
FAQ
What is prokaryotic translation?
Prokaryotic translation is the process by which proteins are synthesized in prokaryotic organisms, such as bacteria. It involves the conversion of the genetic information stored in messenger RNA (mRNA) molecules into functional proteins.
What are the key components involved in prokaryotic translation?
The main components involved in prokaryotic translation are mRNA, ribosomes, transfer RNA (tRNA), amino acids, and various protein factors, including initiation factors, elongation factors, and release factors.
How does prokaryotic translation initiation occur?
In prokaryotes, translation initiation begins with the binding of the small ribosomal subunit to the mRNA molecule. This complex then recognizes a specific start codon, usually AUG, and recruits the initiator tRNA carrying N-formylmethionine (fMet). The large ribosomal subunit joins the complex, forming the initiation complex.
What is the role of tRNA in prokaryotic translation?
tRNA molecules act as adapters between the mRNA and amino acids during protein synthesis. Each tRNA molecule recognizes a specific codon on the mRNA through its anticodon sequence and carries the corresponding amino acid, which is then added to the growing polypeptide chain.
How does prokaryotic translation elongation occur?
During elongation, the ribosome moves along the mRNA in a 5′ to 3′ direction. Each incoming aminoacyl-tRNA enters the ribosome at the A site, where it forms a codon-anticodon base pairing with the mRNA. The ribosome catalyzes the formation of a peptide bond between the amino acid on the A site tRNA and the growing polypeptide chain on the P site tRNA. The ribosome then translocates, shifting the tRNAs to the P and E sites, respectively.
What is the role of ribosomes in prokaryotic translation?
Ribosomes are the cellular organelles responsible for protein synthesis. They consist of two subunits, a small subunit that binds to mRNA and a large subunit that catalyzes peptide bond formation. Ribosomes provide the platform for tRNA binding, mRNA decoding, and the joining of amino acids to form a polypeptide chain.
How is prokaryotic translation terminated?
Translation termination occurs when the ribosome reaches a stop codon (UAA, UAG, or UGA) on the mRNA. Release factors recognize the stop codon and bind to the ribosome, causing the release of the completed polypeptide chain and the dissociation of the ribosomal subunits.
Are there any regulatory mechanisms involved in prokaryotic translation?
Yes, prokaryotic translation can be regulated at various levels. Regulatory proteins can bind to specific sequences on the mRNA, affecting translation initiation or elongation. Additionally, small RNA molecules, called riboswitches, can bind to mRNA and influence translation efficiency based on cellular conditions.
How is prokaryotic translation different from eukaryotic translation?
Prokaryotic translation occurs in the cytoplasm and does not involve extensive RNA processing steps like splicing. Eukaryotic translation occurs in the cytoplasm and can also occur in the endoplasmic reticulum (for membrane-bound proteins). Eukaryotic translation involves more complex initiation and elongation processes and requires additional protein factors.
What are the applications of studying prokaryotic translation?
Understanding prokaryotic translation is important for various areas of research and applications, including the development of antibiotics targeting bacterial translation, the engineering of recombinant proteins in bacteria, and the study of gene expression and regulation in prokaryotes.
References
- Rodnina MV. Translation in Prokaryotes. Cold Spring Harb Perspect Biol. 2018 Sep 4;10(9):a032664. doi: 10.1101/cshperspect.a032664. PMID: 29661790; PMCID: PMC6120702.
- Principles of Genetics, D. Peter Snustad, Michael J. Simmons.
- Biochemistry, Dr. U. Satyanarayana, Dr. U. Chakrapani.
- https://en.wikipedia.org/wiki/Translation_(biology)
- https://www.onlinebiologynotes.com/translation-in-prokaryotes/
- https://www.biologydiscussion.com/cell/prokaryotes/translation-in-prokaryotes-genetics/38022
- https://tmv.ac.in/ematerial/zoology/ks/PGSEM2202Prokaryotictranslation.pdf
- https://www.biologydiscussion.com/cell/prokaryotes/translation-in-prokaryotes-genetics/38022
- https://courses.lumenlearning.com/wm-biology1/chapter/prokaryotic-translation/