What is Primer?
- A primer is a short sequence of nucleotides that is essential for the initiation of DNA synthesis. It functions as the basis for DNA replication or amplification in techniques such as PCR (Polymerase Chain Reaction). Primers are designed to be complementary to particular regions of template DNA, allowing them to bind and serve as a starting point for DNA synthesis.
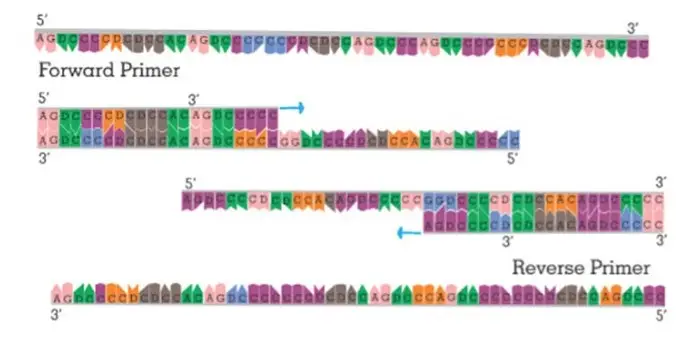
- Typically, a pair of primers consisting of a forward primer and a reverse primer is used in DNA amplification techniques. The forward primer is designed to run in the 3′-5′ direction, whereas the reverse primer is designed to run in the 5′-3′ direction. Together, they bind to opposite extremities of the target sequence, with the 3′ end of the forward primer corresponding to the DNA elongation template strand.
- During DNA elongation, the primers direct the DNA polymerase to produce two new strands of double-stranded DNA. The forward primer guides the synthesis of the new strand in the 5′-3′ direction, whereas the reverse primer guides the synthesis of the complementary strand.
- Nonetheless, the formation of dimers is a potential issue that may arise. When the template strand binds with its complementary strand, an unfavourable condition results in the formation of dimers. In the design of primers, it is essential to avoid such dimers.
- Compared to RNA primers, DNA primers are more commonly used in polymerase chain reaction (PCR). DNA primers are able to withstand the high temperatures involved in the denaturation phase of PCR without degrading. Unlike RNA primers, DNA primers can be readily synthesised and used in the unidirectional mode of polymerization. Primarily DNA primers are utilised in PCR for the amplification of DNA.
- During the replication process, primers have the ability to eliminate RNA primers at the conclusion. However, DNA primers are not eliminated after DNA synthesis or amplification has been completed. Therefore, it is essential to utilise nucleotide primers that target the desired region precisely.

Types of Primers
There are two main types of primers: DNA primers and RNA primers. DNA primers are commonly used in vitro, specifically for PCR amplification and DNA sequencing. On the other hand, RNA primers are utilized in in vivo processes like DNA replication and cloning.
1. DNA Primers:
- Application: Used in vitro for PCR amplification and DNA sequencing.
- Reaction: The amplification process is temperature-dependent and requires fewer proteins.
- Length: Typically 18-24 base pairs.
- Synthesis: DNA primers are chemically synthesized.
- Stability: DNA primers are long-lived and more stable.
2. RNA Primers:
- Application: Primarily used in in vivo processes like DNA replication and cloning.
- Reaction: The replication process is a catalytic reaction that relies on enzymes and involves several proteins.
- Length: Generally 10-20 base pairs.
- Synthesis: RNA primers are synthesized using the Primase enzyme.
- Stability: RNA primers are short-lived and more reactive.
During DNA replication, DNA polymerase initiates the addition of nucleotides to the reactive 3′ (OH) end of the existing nucleic acid, facilitating the elongation and replication of the parent strand. DNA primers are preferred in various applications due to their stability, ease of storage, and the requirement of fewer enzymes to initiate synthesis.
The choice between DNA primers and RNA primers depends on the specific experimental requirements and the nature of the process, whether it is in vitro amplification or in vivo replication and cloning. DNA primers are favored for their longevity and stability, while RNA primers are more short-lived and reactive.
DNA primers vs RNA primers
| Characteristics | DNA Primers | RNA Primers |
|---|---|---|
| Application | In vitro: PCR amplification, DNA sequencing | In vivo: DNA replication, cloning |
| Reaction | Temperature-dependent, fewer proteins involved | Enzyme-dependent, catalytic reaction with several proteins |
| Length | 18-24 base pairs | 10-20 base pairs |
| Synthesis | Chemically synthesized | Synthesized by Primase enzyme |
| Stability | Long-lived and more stable | Short-lived and more reactive |
Primer Designing
Primer designing is a critical process that involves considering several essential factors. Let’s explore these factors and their significance:
- Primer Length: Optimal primer length is typically between 18 to 24 nucleotides. Shorter primers facilitate easy binding to the template at the annealing temperature during PCR.
- Melting Temperature (Tm): Tm refers to the temperature at which half of the primer-template duplexes are dissociated. The GC content of the primer sequence provides an estimate of Tm. The difference in Tm between the forward and reverse primers should not be less than 2°C.
- Primer Annealing (Ta): Ta is the temperature at which primers anneal to the template DNA during PCR. A high Ta can result in insufficient primer-template hybridization and low PCR product yield. Conversely, a low Ta may lead to non-specific PCR products due to a high number of mismatches. Ta can be calculated using the equation: Ta = 0.3 * Tm (primer) + 0.7 * Tm (product) – 14.9.
- Primer GC Content and Clamp: For gene sequencing, primers should ideally have a GC content between 40% and 60%. A GC clamp, consisting of two GC bases at the 3′ end of the primer, enhances primer stability and improves binding specificity. GC base pairs form three hydrogen bonds, which are stronger than the two hydrogen bonds in AT base pairs, contributing to primer stability.
- Setting Restriction Enzyme (RE) Cut Sites: When designing primers for cloning or restriction enzyme digestion, adding 3-5 bases to the 5′ end of the target primer can create a leader sequence that facilitates efficient cutting by the corresponding restriction enzyme.
- End Stability: A stable 3′ end of the primer, often achieved by incorporating a maximum number of G bases, improves primer binding specificity and reduces the chances of false priming.
By considering these factors, researchers can design primers that are optimized for specific applications, such as PCR amplification, gene sequencing, or cloning, leading to successful and specific amplification of the target DNA sequence.
Designing PCR Primers Cautions
When designing PCR primers, certain cautionary aspects need to be considered. Let’s explore these cautionary factors and their significance:
- Hairpins: Hairpin structures are formed by intramolecular interactions within a primer, specifically at the 3′ end. It is generally acceptable if the hairpin structure has a stability of -2 kcal/m, and internal hairpins should have a stability of -3 kcal/m.
- Repeats and Runs: The presence of consecutive dinucleotide repeats or runs within a primer can affect its specificity and efficiency. It is important to limit the occurrence of repeats and runs, with a maximum of 4 dinucleotides and 4 base pairs.
- Dimers: Dimers are formed when two primers interact with each other, resulting in double-stranded DNA. Self-dimers occur when two identical or homologous primers interact, while cross-dimers refer to interactions between forward and reverse primers. Care should be taken to avoid both self-dimer and cross-dimer formation.
- Primer-Template Cross Homology: Primers should be designed to have minimal homology within the template sequence, except for the target site. Intra-primer homology refers to complementary bases within the same primer, with regions longer than 3 bases potentially causing intramolecular bonding. Inter-primer homology occurs when forward and reverse primers have complementary sequences, leading to intermolecular bonding.
- Primer Dimer Formation: It is crucial to analyze and prevent the formation of primer dimers. Primer dimer formation can be assessed by calculating the Gibbs free energy. Analyzing the 5′ end of the primer has been found to be more reliable than the 3′ end.
Considering these cautionary factors helps ensure the specificity, efficiency, and accuracy of PCR amplification. By avoiding hairpins, dimers, excessive repeats and runs, unintended homology, and primer dimer formation, researchers can design primers that specifically target the desired DNA sequence and minimize non-specific binding and amplification.
Primer design Protocol (Steps/Process of Primer design)
Primer design plays a crucial role in various cloning methods such as Golden Gate and Gibson cloning. Let’s explore the primer design protocols for these methods:
Primer Design for Golden Gate Cloning Method:
- Identify DNA Fragments: Determine the DNA fragments to be assembled into a single construct using Golden Gate cloning.
- Overlap Design: Design PCR primers with overlaps that create restriction sites with adjacent DNA fragments. These overlaps are typically 15-30 base pairs long and form non-palindromic sticky end overhangs.
- Type 2S Enzyme Selection: Select a Type 2S restriction enzyme compatible with the chosen vector and DNA fragments. This enzyme allows for the directional assembly of fragments.
- DNA Fragment Amplification: Perform PCR amplification using the designed primers to amplify the DNA fragments of interest.
- Assembly: Mix the PCR-amplified fragments and the linearized vector along with the Type 2S enzyme and DNA ligase. The fragments are directionally assembled into the vector by the sticky end overhangs.
- Cloning: Transform the assembled construct into a suitable host organism for cloning.
Primer Design for Gibson Cloning Method:
- Identify Target DNA Fragments: Determine the target DNA fragments to be assembled using the Gibson cloning method.
- Overlap Design: Design primers that contain 20-40 base pair overlapping regions at each end of the linearized vector and the target DNA fragments. These overlapping regions should be identical and complementary to facilitate recombination.
- PCR Amplification: Perform PCR amplification using the designed primers to generate the linearized vector and the target DNA fragments with overlapping ends.
- Exonuclease Treatment: Treat the PCR products with an exonuclease to remove the overlapping regions, leaving complementary overhangs for assembly.
- Assembly: Mix the linearized vector and the target DNA fragments with the complementary overhangs. The complementary regions facilitate recombination and fusion of the DNA fragments.
- Cloning: Transform the assembled construct into a suitable host organism for cloning and further analysis.
These protocols outline the steps involved in primer design for Golden Gate and Gibson cloning methods. By carefully designing primers with appropriate overlaps and considering the specific requirements of each method, efficient and successful cloning can be achieved.
Primers and Probes in qPCR Validation
Validation of primers and probes in qPCR is an important step to ensure the specificity and reliability of the assay. Here’s a description of the process:
- In silico Validation: After designing qPCR primers and probes using available tools, in silico validation is performed using the BLAST algorithm. BLAST performs a sequence-similarity search against various databases to confirm the specificity of the targeted gene sequences. The query coverage and maximum identity should be 100%. The BLAST program also provides an “expectation value” (E-value) to indicate the likelihood of finding a match by chance. E-values ≤ 0.01 are considered significant, indicating homologous sequences.
- Empirical Validation: While in silico tools provide valuable feedback, the specificity of the qPCR assay using the designed primers and probes should be validated empirically through direct experimental evidence. This involves conducting the qPCR assay using the primers and probes and analyzing the results.
- Melting Curve Analysis: To assess the specificity of the qPCR product and identify any non-specific amplification, melting curve analysis is performed. This analysis is carried out using qPCR protocols based on double-stranded DNA (dsDNA) binding dyes such as SYBR Green. These dyes bind to primer-dimer and other reaction artifacts, producing a fluorescent signal. By analyzing the melting curves, any non-specific amplification can be detected.
- Software Programs: Melting curve analysis can be performed using various software programs available for qPCR analysis. These programs allow the visualization and analysis of the melting curves obtained after the qPCR amplification.
By validating primers and probes in silico and through empirical validation methods, including melting curve analysis, the specificity and reliability of the qPCR assay can be ensured. It is crucial to validate the primers and probes to obtain accurate and meaningful results in qPCR experiments.
Best online tools for Primer design
- Primer3: http://bioinfo.ut.ee/primer3-0.4.0/
- NCBI Primer-BLAST: https://www.ncbi.nlm.nih.gov/tools/primer-blast/
- OligoAnalyzer: https://www.idtdna.com/calc/analyzer
- PrimerQuest (IDT): https://www.idtdna.com/pages/tools/primerquest
- Geneious Prime: https://www.geneious.com/
- Primer Premier: https://www.premierbiosoft.com/primerdesign/index.html
Primer binding site
- The primer binding site refers to the specific region within a nucleotide sequence where a single-stranded RNA or DNA primer binds to initiate the process of replication. This binding site is present on one of the two complementary strands of a double-stranded nucleotide polymer, specifically on the strand that is to be copied. It can also be found within a single-stranded nucleotide polymer sequence.
- During DNA replication, which is a semi-conservative process, the two DNA strands replicate themselves, resulting in two identical copies of DNA. This process is termed semi-conservative because each copy of DNA contains one strand from the original DNA molecule and one strand from the newly-synthesized DNA molecule.
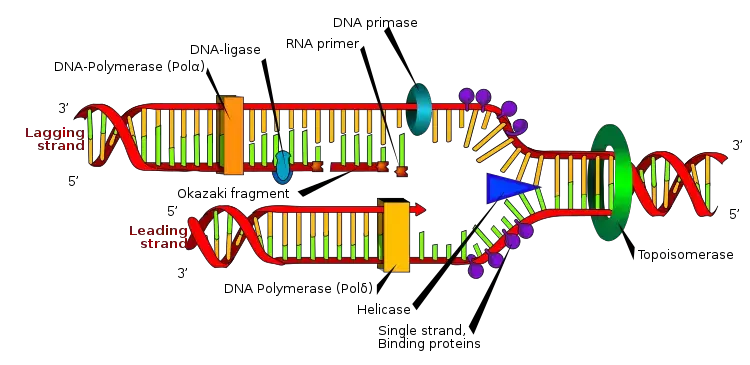
- In DNA replication, an RNA primer is utilized as a short chain of single-stranded RNA. It consists of approximately five to ten nucleotides that are complementary to the DNA template strand. The RNA primer provides the starting point for DNA polymerase to synthesize a new complementary DNA strand to the template strand, but only in the 5′ to 3′ direction.
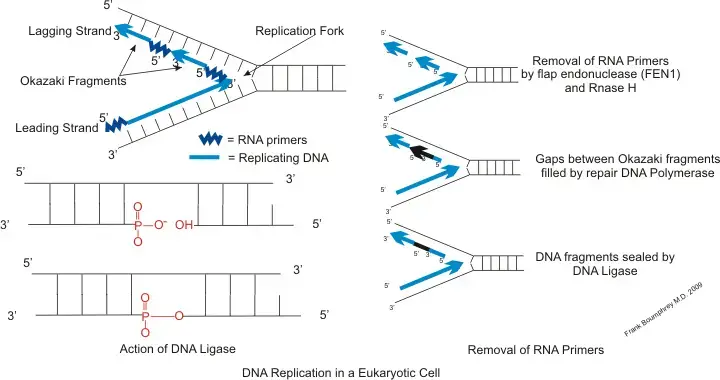
- During replication, one of the newly synthesized strands, known as the leading strand, moves in the 5′ to 3′ direction until it reaches the replication fork. This allows DNA polymerase to utilize the RNA primer and generate a new complementary DNA strand to the template strand.
- On the other hand, the lagging strand moves away from the replication fork in the 3′ to 5′ direction and is composed of small fragments called Okazaki fragments. DNA polymerase synthesizes the lagging strand by employing a new RNA primer for each Okazaki fragment it encounters.
- In summary, the leading strand requires only one RNA primer, while the lagging strand necessitates a new RNA primer for each Okazaki fragment it encounters. The primer binding site plays a crucial role in initiating DNA replication and ensuring accurate and efficient synthesis of new DNA strands.
Dna Primer In Pcr
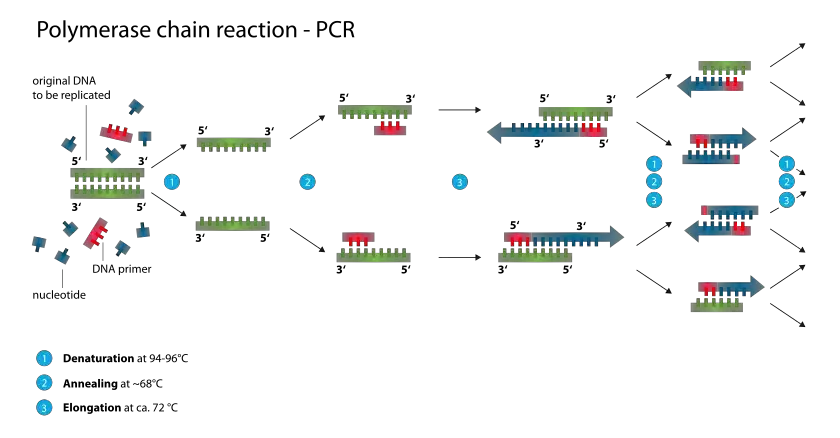
In the context of the polymerase chain reaction (PCR), a DNA primer is a short segment of DNA that plays a crucial role in targeting a specific DNA sequence for amplification. Typically, a DNA primer consists of 18 to 24 base pairs.
The primary purpose of a DNA primer in PCR is to provide a starting point for DNA synthesis by DNA polymerase. During PCR, the DNA template is denatured, separating the two strands. When the temperature is lowered, the DNA primers anneal or bind to their complementary sequences on the template DNA. These primers serve as the initiation points for DNA synthesis.
The binding of the DNA primers to their target sequences allows DNA polymerase to extend the primers by adding complementary nucleotides, leading to the amplification of the specific DNA region of interest. The repetitive cycles of denaturation, primer annealing, and DNA synthesis in PCR result in exponential amplification of the target DNA sequence.
The use of DNA primers in PCR enables the targeted amplification of specific DNA regions for further analysis, such as DNA sequencing or detection of specific genetic markers. By designing primers that specifically bind to the desired target sequence, researchers can selectively amplify and analyze specific regions of the genome.
It is important to note that in living organisms, primers are typically short sequences of RNA. They are synthesized by an enzyme called primase, which is a specialized RNA polymerase. Primers containing RNA are utilized during DNA replication in vivo to initiate the synthesis of new DNA strands.
In summary, DNA primers are short DNA sequences used in PCR to target specific DNA regions for amplification and subsequent analysis. They serve as the starting point for DNA synthesis and are essential for the success of the PCR process.
Types of Primer in PCR
In the polymerase chain reaction (PCR), two types of primers are commonly used: forward primers and reverse primers.
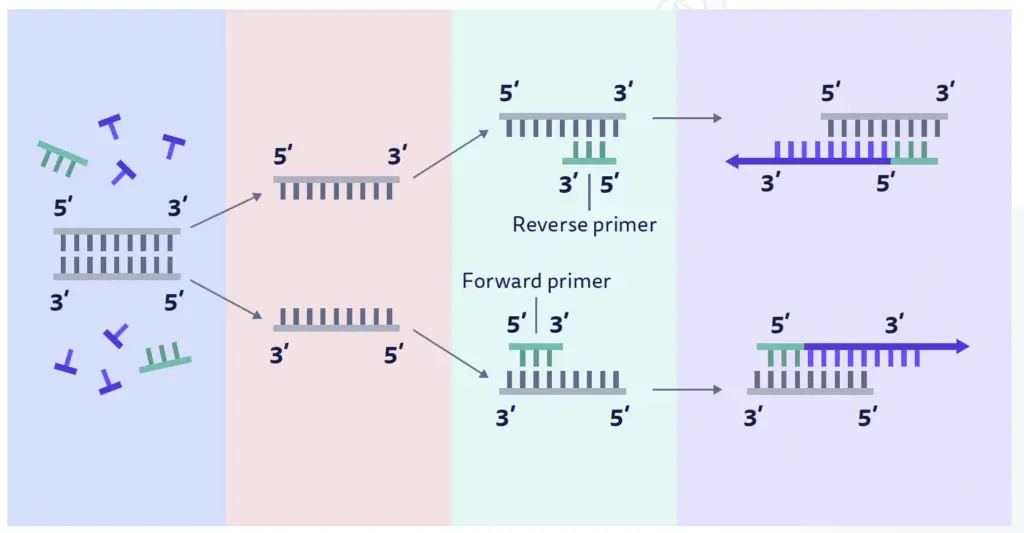
- Forward Primers: Forward primers are designed to anneal to the complementary or negative strand of the double-stranded DNA template. They are typically used to initiate DNA synthesis in the 5′ to 3′ direction. The antisense strand of the DNA acts as the template strand during PCR, and the forward primer binds to this template to start the amplification process. The newly synthesized strand of DNA, which is complementary to the template strand, is also known as the coding strand.
- Reverse Primers: Reverse primers, on the other hand, anneal to the positive strand of the double-stranded DNA template. The positive-sense strand is the opposite strand to the template DNA strand and is also referred to as the anticoding strand. Reverse primers are designed to initiate DNA synthesis in the 3′ to 5′ direction. They bind to the positive strand and facilitate the amplification of the DNA region in the opposite direction compared to forward primers.
In PCR, both forward and reverse primers are used together to amplify the desired DNA region. The forward primer binds to the antisense strand, and the reverse primer binds to the positive-sense strand, flanking the target DNA sequence. This allows DNA polymerase to synthesize new DNA strands in both directions from the primers, resulting in the amplification of the specific DNA region of interest.
The selection and design of appropriate forward and reverse primers are crucial for the success of PCR experiments. They should have complementary sequences to the target DNA region, with optimal melting temperatures and minimal formation of secondary structures or primer-dimer interactions. Proper primer design ensures specific and efficient amplification of the target DNA during PCR.
Applications of Primer Design
Primer design plays a crucial role in various applications in molecular biology and genetic research. Here are some common applications where primer design is essential:
- Polymerase Chain Reaction (PCR): PCR is a widely used technique for amplifying specific DNA sequences. Primer design is crucial for PCR as the primers define the regions to be amplified. Proper primer design ensures specific and efficient amplification of the target DNA.
- Real-time Quantitative PCR (qPCR): qPCR is used for the quantification of gene expression levels, detection of pathogens, and genotyping. Accurate primer design is critical for qPCR to ensure specificity and sensitivity in the detection and quantification of target nucleic acids.
- DNA Sequencing: Primer design is essential in DNA sequencing techniques such as Sanger sequencing and next-generation sequencing (NGS). Primers are used to initiate DNA synthesis and sequence specific regions of interest. Proper primer design ensures accurate and reliable sequencing results.
- Cloning: Primer design is necessary for various cloning techniques such as restriction enzyme-based cloning, Gibson assembly, and Golden Gate assembly. Primers are designed to amplify DNA fragments, add necessary restriction enzyme sites, or create overlaps for fusion of DNA fragments.
- Site-Directed Mutagenesis: Primers are designed for introducing specific mutations into a DNA sequence. These primers contain the desired mutation and are used in mutagenesis techniques to generate specific genetic alterations in a target sequence.
- Genotyping: Primers are designed for genotyping applications such as single nucleotide polymorphism (SNP) analysis or detection of genetic variations. Specific primers can differentiate between different alleles or variations in the target DNA sequence.
- Probe Design: In addition to primers, probe design is crucial for applications such as fluorescence in situ hybridization (FISH), hybridization-based assays, and probe-based genotyping. Probes are designed to hybridize specifically with target sequences and allow for detection or identification.
Proper primer design is essential in these applications to ensure specific amplification, accurate quantification, reliable sequencing, efficient cloning, and accurate genotyping. The design parameters such as primer length, melting temperature, and specificity need to be carefully considered to achieve successful results in these molecular biology techniques.
Limitations
While primer design is a critical step in various molecular biology applications, there are some limitations and challenges associated with it. Here are a few limitations of primer design:
- Sequence Variability: Primer design becomes challenging when targeting regions with high sequence variability, such as regions with frequent mutations or polymorphisms. In such cases, it may be difficult to design primers that can specifically amplify the target region without cross-reactivity or non-specific binding.
- Secondary Structures: Secondary structures like hairpins and dimers can form within primers or between primers and templates. These structures can hinder primer binding and affect PCR efficiency. Careful primer design strategies are required to avoid secondary structure formation.
- Specificity and Off-target Effects: Despite in silico validation, there is a possibility of primers binding to unintended targets in the genome. This can lead to off-target amplification and false results. Experimental validation and optimization are necessary to ensure primer specificity.
- Primer-dimer Formation: Primer-dimer formation occurs when primers anneal to each other and amplify nonspecific products. This can reduce the efficiency and specificity of PCR. Careful primer design and optimization of reaction conditions are necessary to minimize primer-dimer formation.
- Primer Length and GC Content: The length and GC content of primers can affect primer specificity and annealing efficiency. Longer primers may increase specificity but can reduce annealing efficiency, while extreme GC content can affect primer stability and amplification efficiency. Optimal primer length and GC content need to be considered during primer design.
- Target Accessibility: Primers may not effectively bind to target regions that are structurally inaccessible or tightly bound to other proteins or nucleic acids. This can result in inefficient amplification or failure to detect the target sequence.
- Cost and Time: Designing primers can be time-consuming, especially when targeting complex genomes or multiple gene regions. Additionally, the cost associated with primer synthesis and experimental validation should be considered.
Despite these limitations, advancements in primer design tools, optimization strategies, and experimental validation techniques have significantly improved primer design efficiency and specificity. It is important to carefully consider these limitations and optimize primer design parameters to ensure successful and reliable molecular biology experiments.

FAQ
What is a primer in molecular biology?
A primer is a short nucleic acid sequence (DNA or RNA) that is complementary to a specific target sequence. It serves as a starting point for DNA synthesis or amplification during various molecular biology techniques.
Why are primers necessary in PCR?
Primers are necessary in PCR (Polymerase Chain Reaction) to define the region of DNA to be amplified. They bind to the complementary target DNA and provide a starting point for DNA synthesis by DNA polymerase.
How are primers designed?
Primers are designed based on the target DNA sequence using specific criteria such as length, melting temperature (Tm), GC content, and absence of secondary structures or primer-dimer formation. Several online tools are available to aid in primer design.
What is the melting temperature (Tm) of a primer?
The melting temperature (Tm) of a primer is the temperature at which half of the primer molecules are in a single-stranded form and half are in a double-stranded form. It is an important parameter used to determine the optimal annealing temperature for PCR or other nucleic acid hybridization techniques.
Can the same primer be used for different DNA templates?
In some cases, a primer designed for a specific target sequence can be used for different DNA templates that share the same target region. However, careful consideration should be given to sequence variations and potential off-target binding.
What is the ideal length for a primer?
In general, primers are typically 18-24 nucleotides long for PCR amplification. However, the optimal primer length may vary depending on the specific application and target sequence.
Can RNA primers be used in PCR?
No, PCR primarily utilizes DNA primers. RNA primers are typically involved in DNA replication and other in vivo processes, whereas in vitro techniques like PCR rely on DNA primers for amplification.
How can primer specificity be ensured?
Primer specificity can be ensured by performing in silico validation, such as BLAST analysis, to check for potential cross-reactivity or non-specific binding. Experimental validation, including optimization of reaction conditions and analysis of melting curves, is also crucial to confirm primer specificity.
What are the common challenges in primer design?
Some common challenges in primer design include dealing with high sequence variability, avoiding secondary structure formation, ensuring specificity, minimizing primer-dimer formation, and optimizing primer length and GC content.
Are commercial primer sets available for specific applications?
Yes, many commercial suppliers offer pre-designed primer sets for specific applications, such as gene expression analysis, genotyping, or pathogen detection. These primer sets are designed and validated by experts, providing convenience and reliability for researchers.
References
- Griep, M. A. (2001). Primer RNA. Encyclopedia of Genetics, 1546–1548. doi:10.1006/rwgn.2001.1025
- Griep, M. A. (2017). Primer RNA ☆. Reference Module in Life Sciences. doi:10.1016/b978-0-12-809633-8.06952-1
- https://pharmaceuticalintelligence.com/2014/07/29/a_primer_on_dna_and_dna_replication/
- https://www.news-medical.net/life-sciences/What-is-DNA-Polymerase.aspx
- https://www.news-medical.net/life-sciences/Mechanism-of-DNA-Synthesis.aspx
- https://www.genome.gov/genetics-glossary/Primer
- https://www.addgene.org/protocols/primer-design/
- https://www.benchling.com/primers-in-dna-replication
- https://geneticeducation.co.in/comparison-between-dna-primer-and-rna-primer/
- https://www.chegg.com/learn/topic/dna-primer-in-pcr
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/biotechnology/a/polymerase-chain-reaction-pcr
- https://www.the-odin.com/primer-design/
- http://bioweb.uwlax.edu/GenWeb/Molecular/seq_anal/primer_design/primer_design.htm
- https://www.sciencedirect.com/science/article/pii/S2211124717314869
- https://en.wikipedia.org/wiki/Primer_binding_site