What is Point mutation?
- A point mutation is a specific type of genetic mutation that involves a change in a single nucleotide base within the DNA or RNA sequence. This alteration can manifest as a substitution, insertion, or deletion of one base pair. In the DNA sequence, point mutations can involve the nitrogenous bases cytosine (C), guanine (G), adenine (A), and thymine (T), whereas in RNA, uracil (U) replaces thymine.
- Point mutations generally occur due to errors during DNA replication, though they can also be induced by external factors such as X-rays or ultraviolet radiation. The primary impact of point mutations on the genetic code involves the alteration of the encoded protein’s amino acid sequence. Since nucleotides are read in triplets during translation, a single base change can lead to significant variations in protein structure and function.
- The effects of point mutations vary widely depending on their specific nature. Some point mutations may be neutral, causing no observable changes in protein function. Others can be deleterious, potentially disrupting protein production or function. For example, missense mutations, a subtype of point mutations, result in the incorporation of an incorrect amino acid, which may affect the protein’s activity. Conversely, nonsense mutations introduce premature stop codons, leading to truncated and often nonfunctional proteins.
- Overall, the severity and consequences of a point mutation are influenced by its location within the gene and the role of the affected protein. Therefore, while some point mutations may have minimal impact, others can significantly affect cellular processes and contribute to various genetic disorders.
Definition of Point mutation
A point mutation is a genetic mutation involving a change in a single nucleotide base in the DNA or RNA sequence, which can affect the sequence of amino acids in a protein.
Causes of Point mutation
Point mutations are primarily caused by various mechanisms that affect the DNA sequence. These mutations can occur during DNA replication or due to external factors. Here are the key causes of point mutations:
- DNA Replication Errors: Point mutations often arise during DNA replication. During this process, the DNA molecule is copied, and errors can occur if incorrect nucleotides are incorporated into the new strand. Even a single incorrect nucleotide can alter the entire DNA sequence.
- Spontaneous Mutations: Some point mutations occur spontaneously due to natural errors in cellular processes. These errors are random and can happen without external influence, often resulting from the inherent instability of DNA.
- Mutagens: Exposure to mutagens significantly increases the rate of point mutations. Mutagens are agents that induce mutations and can be physical or chemical:
- Physical Mutagens: Include radiation such as ultraviolet (UV) light and X-rays. UV light can cause thymine dimers, which disrupt DNA structure. X-rays can lead to ionization of DNA molecules, causing breaks and changes in the sequence.
- Chemical Mutagens: Include substances that chemically alter DNA bases. These chemicals can mispair nucleotides or disrupt the helical structure of DNA, leading to incorrect base pairing during replication.
- Reactive Oxygen Species: Byproducts of cellular metabolism, such as free radicals, can damage DNA. These reactive oxygen molecules can cause oxidative stress, leading to base modifications or breaks in the DNA strand.
- Degradation of DNA Bonds: Over time, DNA bonds can degrade due to various factors, including environmental conditions. This degradation can result in mutations if the DNA is not repaired properly.
- Nucleotide Substitutions: Errors during DNA replication or repair processes can lead to nucleotide substitutions, where one base is replaced by another. This change can alter the codon sequence and potentially affect protein function.
- Insertions and Deletions: Point mutations can also occur through the insertion or deletion of nucleotides. These changes can disrupt the reading frame and lead to significant alterations in the resulting protein.
- Errors in DNA Repair: Mistakes during DNA repair processes can introduce point mutations. If the repair machinery fails to accurately correct damaged DNA, mutations can persist in the genome.
- Aging: As cells age, the DNA replication and repair mechanisms may become less efficient, increasing the likelihood of point mutations over time.
- Environmental Factors: Exposure to various environmental factors, such as pollutants or chemicals, can contribute to the induction of point mutations by interacting with the DNA and causing structural changes.
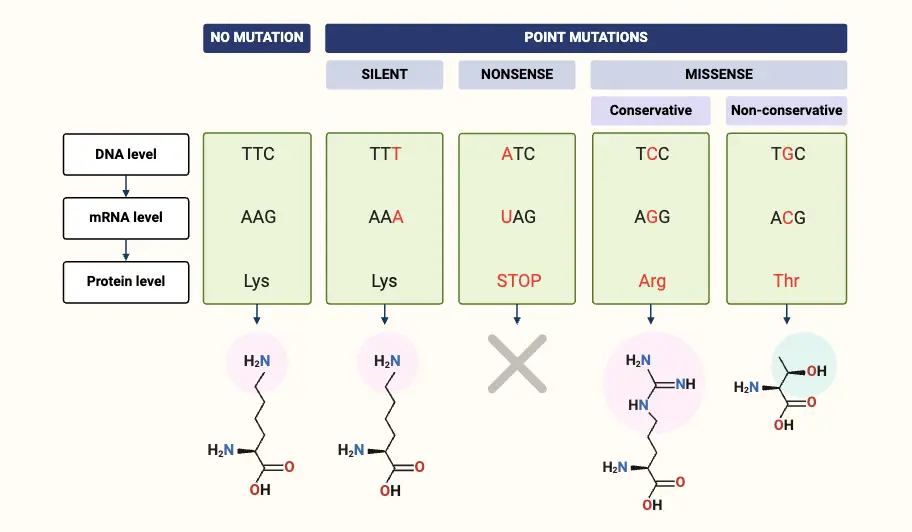
Types of Point mutation
Point mutations are categorized based on the nature of the changes they induce in the DNA sequence and their subsequent effects on protein function. These mutations can be broadly classified into different types, each with distinct implications.
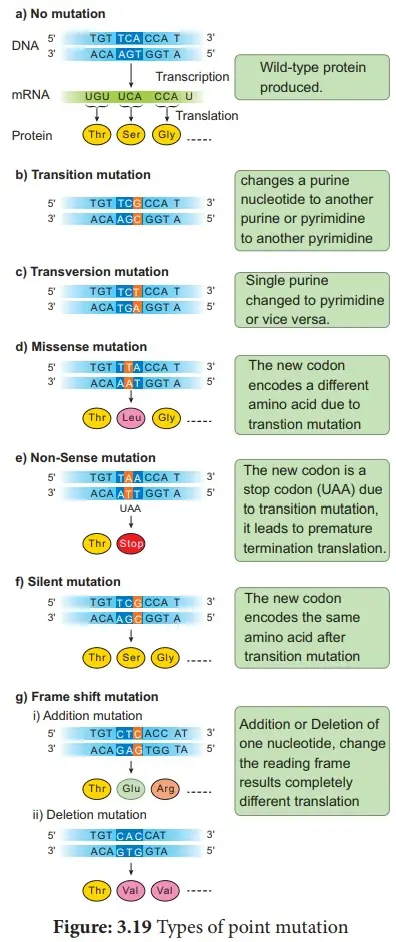
- Transition Mutations
- Definition: Transition mutations occur when a purine base is substituted for another purine base or a pyrimidine base is replaced by another pyrimidine base.
- Characteristics: These mutations involve changes between the two types of nitrogenous bases, maintaining the same chemical class. For instance, adenine (A) may be replaced by guanine (G) or cytosine (C) by thymine (T).
- Examples: An example includes the replacement of adenine with guanine. Transition mutations are more common due to the simpler nature of the chemical changes involved.
- Implications: They are less likely to cause significant disruptions in protein function, often resulting in silent mutations if the amino acid sequence remains unchanged.
- Transversion Mutations
- Definition: Transversion mutations involve the substitution of a purine base with a pyrimidine base or vice versa.
- Characteristics: This type of mutation represents a more drastic change compared to transition mutations because it involves the replacement of bases with different structural forms—purines (adenine and guanine) with pyrimidines (cytosine and thymine).
- Examples: An example includes the substitution of adenine with cytosine or thymine with guanine. There are eight possible transversion changes.
- Implications: Transversions are less frequent but can have more pronounced effects on the protein, often leading to significant alterations in protein structure and function.
- Nonsense Mutations
- Definition: Nonsense mutations occur when a base substitution results in the formation of a premature stop codon.
- Characteristics: This type of mutation changes a codon that originally specified an amino acid into one of the three stop codons (UAG, UAA, UGA), leading to early termination of protein synthesis.
- Examples: In beta-thalassemia, a nonsense mutation in the gene coding for beta-globin creates a premature stop codon, producing a truncated and nonfunctional hemoglobin protein.
- Implications: Nonsense mutations typically result in nonfunctional proteins and can cause severe genetic disorders due to the loss of essential protein function.
- Missense Mutations
- Definition: Missense mutations involve the substitution of a single nucleotide that changes one amino acid in the protein sequence.
- Characteristics: This type of mutation can lead to the incorporation of a different amino acid into the protein, potentially altering its structure and function.
- Examples: Sickle cell anemia is caused by a missense mutation where glutamic acid is replaced by valine in the hemoglobin protein, affecting the red blood cell structure and function.
- Implications: Missense mutations can result in proteins with altered functions, which may be benign, neutral, or deleterious depending on the nature of the amino acid change and its impact on the protein’s activity.
- Silent Mutations
- Definition: Silent mutations are point mutations that do not alter the amino acid sequence of the protein.
- Characteristics: These mutations occur when a change in the nucleotide sequence does not affect the final amino acid produced due to the redundancy in the genetic code.
- Examples: Replacing thymine with cytosine in a codon that still codes for the same amino acid (e.g., TTC to TTT, both coding for lysine).
- Implications: Silent mutations generally have no impact on protein function or phenotype but can affect the stability or expression of mRNA.
- Frameshift Mutations
- Definition: Frameshift mutations occur when nucleotides are inserted into or deleted from the DNA sequence, causing a shift in the reading frame of the codons.
- Characteristics: This type of mutation alters the downstream codon sequence, leading to a completely different amino acid sequence from the point of mutation onwards.
- Examples: In Crohn’s disease, an insertion mutation causes a shift in the reading frame of the NOD2 gene, resulting in a truncated protein.
- Implications: Frameshift mutations usually result in significant changes to the protein, often producing nonfunctional proteins and severe phenotypic consequences.
Examples of Point mutation
Point mutations, which involve changes in a single nucleotide base, can lead to a variety of genetic disorders and conditions. Here are some notable examples:
- Sickle Cell Anemia
- Description: Sickle cell anemia is an autosomal recessive disorder caused by a point mutation in the HBB gene on chromosome 11.
- Mutation: A single base substitution changes adenine (A) to thymine (T) in the codon GAG, converting it to GTG. This alters the amino acid sequence, replacing glutamic acid with valine.
- Consequences: The resultant mutant allele, known as HBS, leads to the production of abnormal hemoglobin. This causes red blood cells to adopt a sickle shape, reducing their oxygen-carrying capacity and leading to various health complications.
- Prevalence: This mutation is particularly common in African and Indian populations. Individuals with the homozygous recessive genotype (HBS/HBS) exhibit symptoms, while heterozygous carriers (HBS/normal) generally remain asymptomatic but can pass the mutation to offspring.
- Cystic Fibrosis
- Description: Cystic fibrosis is another autosomal recessive disorder, resulting from mutations in the CFTR gene.
- Mutation: A deletion of three nucleotides (CTT) at position 508 in the CFTR gene, known as ΔF508, leads to the loss of the amino acid phenylalanine. This deletion disrupts the normal function of the CFTR protein.
- Consequences: The mutation results in the production of a defective CFTR protein, which impairs chloride ion transport. This causes thick, sticky mucus to accumulate in various organs, particularly the lungs and pancreas, leading to severe respiratory and digestive issues.
- Neurofibromatosis
- Description: Neurofibromatosis, a genetic disorder characterized by the development of tumors on nerve tissues, can also be caused by point mutations.
- Mutation: Mutations in the NF1 or NF2 genes lead to the production of abnormal neurofibromin or merlin proteins, respectively.
- Consequences: These mutations disrupt normal cell growth and division, leading to tumor formation. The severity of the condition varies depending on the specific mutation and its impact on protein function.
- Tay-Sachs Disease
- Description: Tay-Sachs disease is a fatal autosomal recessive disorder caused by a point mutation in the HEXA gene.
- Mutation: A common mutation involves a single nucleotide change that leads to a defective enzyme responsible for breaking down certain lipids in the brain.
- Consequences: The absence of functional enzyme activity results in the accumulation of lipids, causing progressive neurological damage and, ultimately, early death.
- Color Blindness
- Description: Color blindness, particularly red-green color blindness, is often due to point mutations in the genes encoding color-detecting pigments in the retina.
- Mutation: Mutations in the OPN1LW or OPN1MW genes, which encode for long-wavelength or middle-wavelength photopigments, can impair color discrimination.
- Consequences: Individuals with these mutations experience difficulty distinguishing between certain colors, affecting daily activities and color perception.
- Cancer
- Description: Point mutations are frequently implicated in the development of cancer through their effects on tumor suppressor genes and proto-oncogenes.
- Mutation: Mutations can lead to the inactivation of tumor suppressor genes or the activation of oncogenes. For instance, mutations in the TP53 gene can result in a nonfunctional p53 protein, a key regulator of the cell cycle.
- Consequences: These mutations disrupt normal cell growth and division, contributing to uncontrolled cell proliferation and tumor formation.
Detection of Point Mutations
Detecting point mutations is essential for understanding genetic variations that impact health, development, and disease. Several methodologies are employed to identify these single-base alterations, each with its specific applications and advantages. Here is an overview of the primary techniques used to detect point mutations:
- Polymerase Chain Reaction (PCR):
- Principle: PCR amplifies specific DNA sequences using sequence-specific primers, allowing for the detection of mutations within targeted regions.
- Application: Point mutations can be identified by using primers designed to amplify either the normal or mutant allele. For example, in sickle cell anemia, allele-specific PCR can differentiate between the normal and mutant alleles by employing two distinct sets of primers.
- Advantages: This method is highly sensitive and specific, making it suitable for detecting known point mutations and small-scale genetic variations.
- Allele-Specific PCR:
- Principle: A variation of traditional PCR, allele-specific PCR uses primers that are complementary to either the wild-type or mutant allele.
- Application: By designing primers that hybridize specifically to either the normal or mutated sequence, this technique can effectively differentiate between alleles based on the presence or absence of amplification.
- Advantages: It allows for rapid and precise detection of specific point mutations, particularly useful for clinical diagnostics.
- DNA Sequencing:
- Principle: DNA sequencing determines the exact nucleotide sequence of a DNA segment, allowing for the identification of any single nucleotide variations.
- Application: Sequencing can detect both known and unknown point mutations by comparing the obtained sequence with a reference sequence. Techniques like Sanger sequencing or next-generation sequencing (NGS) are commonly used.
- Advantages: This method provides comprehensive data on genetic variations, including point mutations, and can identify novel mutations that may not be previously characterized.
- Restriction Fragment Length Polymorphism (RFLP):
- Principle: RFLP involves digesting DNA with restriction enzymes that cut the DNA at specific sequences. Variations in the DNA sequence, including point mutations, can alter the cutting pattern of these enzymes.
- Application: By comparing the DNA fragment lengths after restriction enzyme digestion, one can infer the presence of point mutations if they affect the restriction sites.
- Advantages: RFLP is useful for detecting mutations that alter restriction enzyme recognition sites and can provide insights into genetic diversity.
- Fluorescent Dye-Based Methods:
- Principle: This approach uses fluorescent dyes to label DNA fragments, which are then analyzed using specialized equipment to detect sequence variations.
- Application: Fluorescent dyes can be incorporated into sequencing reactions or used in PCR assays to visualize and quantify specific DNA sequences.
- Advantages: The use of fluorescent dyes enhances the sensitivity and accuracy of mutation detection, especially in high-throughput analyses.
- Allele-Specific Hybridization:
- Principle: This method involves hybridizing DNA samples with probes that are specific for either the wild-type or mutant allele.
- Application: Probes are designed to bind selectively to either the normal or mutant allele, allowing for the detection of point mutations through differences in hybridization patterns.
- Advantages: This technique is useful for high-throughput screening of known mutations and can be adapted for large-scale studies.
- Real-Time PCR (qPCR):
- Principle: Real-time PCR quantifies DNA amplification in real-time using fluorescent markers, enabling the detection of specific point mutations during the amplification process.
- Application: By using fluorescent probes or dyes, real-time PCR can monitor the presence of point mutations as the PCR progresses.
- Advantages: This method provides quantitative data and allows for the rapid detection of mutations, suitable for both research and clinical applications.
- Comparative Genomic Hybridization (CGH):
- Principle: CGH compares the DNA of a test sample with a reference sample to identify variations, including point mutations, through differences in hybridization.
- Application: CGH can detect copy number variations and other genetic alterations, including point mutations, by comparing fluorescence intensity.
- Advantages: This technique is effective for detecting genetic abnormalities on a genomic scale.
- Single-Nucleotide Polymorphism (SNP) Genotyping:
- Principle: SNP genotyping involves analyzing specific single nucleotide variations across the genome using various technologies.
- Application: Techniques such as SNP microarrays or high-throughput sequencing can identify and quantify SNPs associated with point mutations.
- Advantages: SNP genotyping provides a comprehensive view of genetic variations and is widely used in genetic studies and personalized medicine.
- Molecular Beacon Assays:
- Principle: Molecular beacons are fluorescent probes that change their fluorescence signal upon hybridization with target sequences, allowing for specific detection of point mutations.
- Application: These assays are used in real-time PCR to detect specific nucleotide changes by monitoring fluorescence changes during amplification.
- Advantages: Molecular beacon assays offer high sensitivity and specificity for detecting point mutations in various genetic contexts.
Applications of Point mutation
The following points outline the key applications of point mutations:
- Molecular Therapy:
- Point mutations are employed in molecular therapy to target and correct genetic defects associated with specific diseases. By introducing precise mutations, scientists can potentially rectify genetic anomalies that contribute to conditions such as cystic fibrosis and muscular dystrophy.
- Cancer Treatment:
- In oncology, point mutations are strategically used to target cancer cells. Mutagens can be utilized to induce specific point mutations that disrupt the function of genes involved in cancer progression. This targeted approach helps in the removal or inactivation of harmful nucleotides, potentially halting cancer growth.
- Mutational Breeding:
- In agriculture, point mutations are harnessed through mutational breeding to enhance crop traits. By inducing point mutations in plant genomes, researchers can develop new crop varieties with improved characteristics, such as higher yields, disease resistance, or better nutritional profiles.
- Genetic Enhancement:
- Point mutations are used to introduce beneficial traits into organisms through genetic engineering. This can involve the development of genetically modified organisms (GMOs) with enhanced abilities or desirable traits, which can be useful in various fields including agriculture, pharmaceuticals, and industrial biotechnology.
- Research and Functional Studies:
- Point mutations are also instrumental in research to study gene function and the impact of specific nucleotide changes on protein structure and function. By creating and analyzing point mutations, researchers can gain insights into the roles of different genes and the mechanisms underlying various diseases.
- Diagnostic Tools:
- Point mutations can serve as markers for diagnosing genetic disorders. Identifying specific point mutations associated with certain diseases allows for accurate genetic testing and early diagnosis, which is crucial for managing and treating genetic conditions.
- Drug Development:
- In drug development, point mutations are utilized to understand drug resistance mechanisms. By studying how point mutations affect drug interactions, scientists can design more effective medications and strategies to overcome resistance in pathogens or cancer cells.
- Biotechnology Applications:
- Point mutations are used in biotechnology to create engineered enzymes with improved properties. By altering specific amino acids through point mutations, researchers can design enzymes with enhanced stability, activity, or specificity for various industrial applications.
- Evolutionary Studies:
- Point mutations provide valuable information for evolutionary biology. By studying point mutations in different species, scientists can trace evolutionary changes and understand how genetic variations contribute to adaptation and species diversity.
- Gene Editing Technologies:
- Advanced gene editing techniques, such as CRISPR-Cas9, often utilize point mutations to create specific genetic modifications. This technology allows for precise editing of DNA sequences to introduce or correct point mutations, facilitating research and therapeutic applications.
Learn
[flashcard id=”58578″]
Facts
- Did you know that point mutations can lead to a single nucleotide change in the DNA sequence, potentially altering the encoded protein’s structure and function?
- Have you heard that substitution mutations, a type of point mutation, involve replacing one base pair with another, which can result in silent, missense, or nonsense mutations?
- Are you aware that nonsense mutations create a premature stop codon in the protein sequence, leading to truncated proteins that are often nonfunctional?
- Can you believe that missense mutations, where one amino acid is replaced by another, can result in proteins with altered functions or stability, affecting various biological processes?
- Did you know that silent mutations, despite changing the nucleotide sequence, do not alter the amino acid sequence of the protein due to the redundancy in the genetic code?
- Have you heard that insertion and deletion mutations, often grouped together as frameshift mutations, can dramatically change the reading frame of the genetic code, leading to significant alterations in protein structure?
- Are you aware that frameshift mutations caused by insertions or deletions can result in proteins that are either shortened, elongated, or completely nonfunctional?
- Can you believe that point mutations can be induced intentionally using mutagens in laboratory settings to study gene function, develop new crop varieties, or create genetically modified organisms?
- Did you know that certain genetic diseases, such as sickle cell anemia and cystic fibrosis, are caused by specific point mutations that disrupt normal protein function and lead to disease symptoms?
- Have you heard that understanding point mutations is essential for advancements in personalized medicine, as it allows for the development of targeted therapies based on an individual’s unique genetic makeup?
- https://biologydictionary.net/point-mutation/
- https://www.brainkart.com/article/Types-of-point-mutations_38228/
- https://geneticeducation.co.in/what-is-a-point-mutation/#Causes_of_point_mutation
- https://en.wikipedia.org/wiki/Point_mutation