What are plasmids?
- Plasmid can be define as small, circular DNA molecule found mainly in bacteria, sometimes also in yeast or archaea.
- It exist independently from the chromosomal DNA, but occasionally it integrate by the chromosome under some condition.
- The size of plasmids are very variable, usually range from few thousand base pair (kbp) to several hundred kbp, depend by the species.
- They carry extra genes which not essential for survival, but often give advantage like antibiotic resistance, metabolic activity, or virulence factors.
- Replication of plasmid occurs autonomously, it’s controlled by own origin of replication (Ori site).
- In bacterial cells, plasmids are transmitted by process like conjugation, transformation, or transduction, etc.
- Some plasmid known as R-plasmid, which carry resistance genes against multiple drugs / antibiotics.
- F-plasmid (fertility factor) is responsible for conjugation in E.coli, it help in DNA transfer between donor and recipient cell.
- Plasmids are used in biotechnology as vectors for cloning genes, producing recombinant proteins, or gene therapy.
- Structure usually consist of origin of replication, selectable marker (like ampR gene), and multiple cloning site (MCS).
- It is also used for genetic engineering, because manipulation is easier and fast, though sometimes stability issue occur during large insertions.
- Some are low-copy while others high-copy number plasmids, depending how many copies stay inside a cell.
- By molecular biology viewpoint, plasmids play crucial role in horizontal gene transfer, helping bacteria adapt quickly by environment.
Copy Number and Physical Nature of Plasmids
- Copy Number of a plasmid can define as the average number of plasmid molecules present per bacterial cell.
- It is usually classified as low-copy (about 1–5 copies) or high-copy (tens to hundreds copies), and intermediate types are also observed.
- Copy number regulation is effected by the plasmid’s origin of replication (Ori) and by plasmid-encoded control elements (antisense RNA, iterons), thus tight or loose control can occur depending on plasmid.
- Low-copy plasmids are often provided with partitioning systems (Par proteins), so during cell division at least one copy is allocated to each daughter cell.
- High-copy plasmids are replicated more frequently, they are maintained by statistical distribution rather than active partitioning, and so stability is achieved by large numbers.
- Copy number is influenced by host strain, growth conditions, metabolic burden, and plasmid size, larger plasmids tend to be kept at lower copy, small ones at higher copy.
- Some experimental manipulations are used to alter copy number (mutation of control region, use of helper plasmid), and these strategies are used in cloning to increase yield of recombinant product.
- Physical Nature of plasmids is most commonly circular double-stranded DNA (dsDNA), though linear plasmids are reported in organisms like Streptomyces and Borrelia.
- In vivo plasmids are usually supercoiled (negatively supercoiled), but during extraction they may appear as nicked-open circles, relaxed circles, and linear forms depending on handling.
- Plasmids are often small (a few kbp) or large (hundreds kbp), and their conformation, size and copy number determine behaviour in electrophoresis or density-gradient centrifugation.
- Conjugative plasmids are characterized by tra genes and transfer (mating pair formation), while non-conjugative plasmids lack transfer machinery and rely on mobilization by helpers.
- Plasmid molecules are described by physical parameters like molecular weight, buoyant density (CsCl), and degree of supercoiling, these are measured experimentally for characterization.
- Some plasmids contain linear ends with proteins attached, others are covalently closed circular DNA, and occasionally concatemers or multimers are seen during replication.
- Stability of plasmid in host is affected by partitioning, addiction systems (toxin-antitoxin), metabolic cost of plasmid-carried genes, temperature and host mutations, thus loss can occur if selection is not maintained.
- By ecology viewpoint, plasmids are important vectors of horizontal gene transfer, and they are instrumental in spread of traits like antibiotic resistance, though efforts are made to prevail (prevail used intentionally) against spread.
Properties of Plasmids
- Plasmids are small, extra-chromosomal DNA molecules, usually circular and double-stranded (dsDNA).
- They are self-replicating, controlled by their own origin of replication (Ori), sometimes independent from main chromosome.
- Exist mainly in bacteria, but also observed in yeast, archaea and few protozoa.
- Can be transferred between cells by conjugation, transformation, or transduction etc, which makes them vital for gene exchange.
- Copy Number vary by plasmid type—some high-copy (many per cell), some low-copy (just one or few copies).
- Carry genes that often provide selective advantages, like resistance against antibiotics / heavy metals / toxins.
- Structure usually circular, but few species show linear form (e.g., Streptomyces).
- Possess supercoiled configuration inside cells, though may become nicked-open or relaxed during extraction.
- Some plasmids are conjugative (carry tra genes), enabling transfer, others non-conjugative but can mobilized with help of others.
- Size ranges from about 1–2 kbp to more than 400 kbp; smaller plasmids are often maintained in large number.
- Stability depends by partition system, toxin–antitoxin modules, and selective pressure maintained by environment.
- Can integrate temporarily with host genome, forming episomes, then excise again.
- Used as vectors in molecular biology / genetic engineering because of their easy manipulation and high yield.
- Contain selectable marker (like ampR, tetR) and cloning site region (MCS) for gene insertion.
- Physical nature influenced by ionic strength, temperature, and enzyme activity (like DNase, topoisomerase), which change conformation.
- By natural function, plasmids involved in many processes—fertility (F plasmid), resistance (R plasmid), colicin production (Col plasmid), etc.
- Some plasmids have addiction systems (post-segregational killing), ensuring retention by killing cells which lose them.
- They act as vectors of horizontal gene transfer, spreading traits rapidly by bacterial population.
- Due to wide diversity and flexibility, plasmids are considered as one of most dynamic elements in microbial evolution.
- In biotechnology, they are exploited for cloning, expression, and therapeutic purpose, though instability sometimes prevail (used wrong intentionally) their success in large-scale cultures.
Plasmid Replication Mechanism
- During cell division, bacterial plasmid replication is independent of its nuclear genome replication, with lengthy pauses occurring between replication events.
- The exact amount of plasmid copies depends on the plasmid type, host organism, and growth conditions.
- Unintended deviations from the regular number of copies are adjusted. There do exist dominant and recessive copy mutants of the wild type.
- Rolling circle, Col E1 type, and Iteron contain replication are the three kinds of plasmid replication.
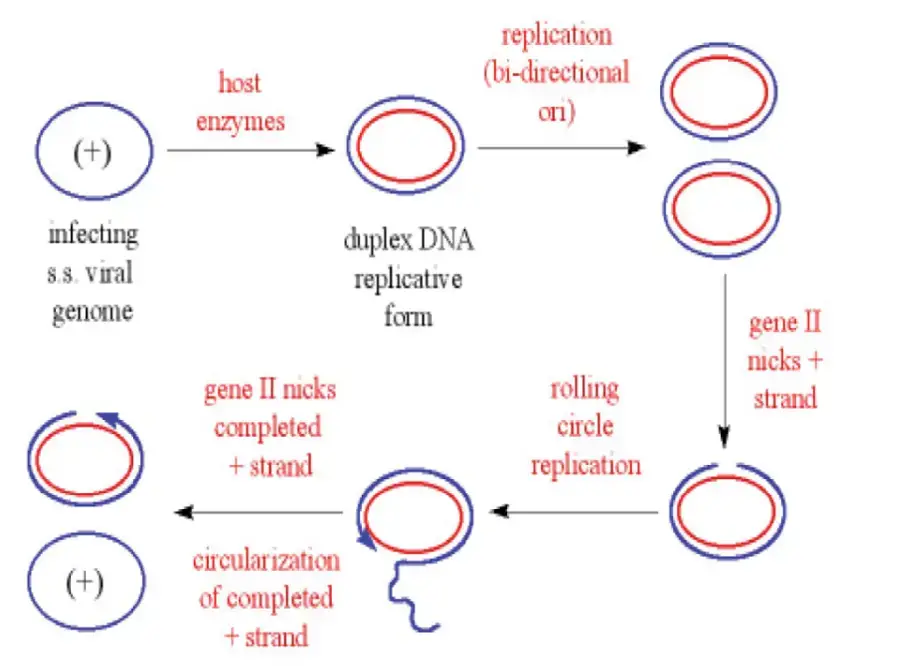
1. Rolling circle Mode of Replication
Rolling-Circle Replication (RCR) is described as a method of DNA replication that is used by some viruses, plasmids and circular genetic elements.
A single-strand nick is introduced at the origin and the 5′-end is displaced, while the 3′-OH end is extended by DNA polymerase.
The replication fork is not formed like in bidirectional replication, instead synthesis is continuous around the circle, and a long single-stranded tail is produced.
The displaced strand is often converted to double-stranded DNA by synthesis primed by short RNAs or hairpins, or it is circularized then used as template.
Host enzymes or virus-encoded proteins are used, depending on organism, so replication proteins may vary by species.
In bacteriophage ΦX174 and many small plasmids RCR is well-documented, and it is favored for rapid copy-number increase.
The nicking enzyme (replicase/initiation protein) is usually sequence-specific, it cleaves at origin then it covalently links to the 5′ end, preventing loss of the strand.
Multiple genome copies can be produced from one circle because the polymerase keeps rolling, thus concatemeric or tandem repeats are generated, then they are processed.
After rolling, cleavage and ligation steps are performed to release unit-length genomes, often by the same nuclease that initiated replication.
RCR is efficient for small circular DNAs, it may prevail (malapropism for “prevent” intended) strand loss when compared with other methods.
The process is simple in principle, yet it is versatile biologically, being adapted by plasmids (rolling plasmid replication), viruses, and some transposable elements.
Procedure of Rolling circle replication in plasmid
- The process start with a covalently closed circular dsDNA molecule, usually of a plasmid that replicate by rolling-circle mode.
- A plasmid-encoded initiator protein (Rep) or nickase enzyme introduces a site-specific nick in one strand (the plus strand) at the double-stranded origin (dso).
- This nick create a free 3′-OH end and a 5′-phosphate end, which are essential for next step of synthesis.
- The Rep protein generally stays bound with the 5′-phosphate end while the free 3′-OH act as a primer for DNA polymerase III.
- Host DNA polymerase III starts synthesizing a new strand using the intact strand as template, displacing the nicked one continuously.
- The displaced single-stranded DNA (ssDNA) moves away from the circle as synthesis proceeds around the molecule.
- In some cases, like Staphylococcus aureus plasmids, a helicase (PcrA) assists in unwinding and displacing the parental strand properly.
- Continuous synthesis may produce several copies of ssDNA in a head-to-tail concatemer, which later are processed individually.
- When the polymerase reaches the original dso, the initiator protein performs another nick, which terminates replication of leading strand.
- This step releases one new dsDNA molecule and one ssDNA intermediate, both are circularized later.
- In plasmids like pT181, the active RepC dimer becomes inactive heterodimer (RepC/C*) after replication to ensure only one round of replication occur.
- The resultant ssDNA now undergoes second phase replication using the single-stranded origin (sso) as initiation site.
- Host RNA polymerase transcribes primer RNA at sso, which serves as primer for complementary strand synthesis.
- DNA polymerase III extends this primer, producing the complementary (lagging) strand and converting ssDNA to dsDNA circle.
- Later, DNA polymerase I removes the RNA primer, fills the space with DNA, and DNA ligase seals the nicks to close the molecule.
- The final dsDNA plasmid is then supercoiled by DNA gyrase to achieve its stable, compact configuration within cell.
- The whole cycle is simple yet smart, allowing plasmid to replicate fastly without complex fork structure, a kind of biological shortcut which prevail extra energy loss.
2. Col E1 type replication
Col E1 type replication is a mode of plasmid replication mainly found in small circular plasmids like ColE1, pBR322, etc.
It can defined as a unidirectional theta-type replication, not by rolling-circle method.
The initiation occur at specific site called origin of replication (oriV), located in region that contain several control sequences.
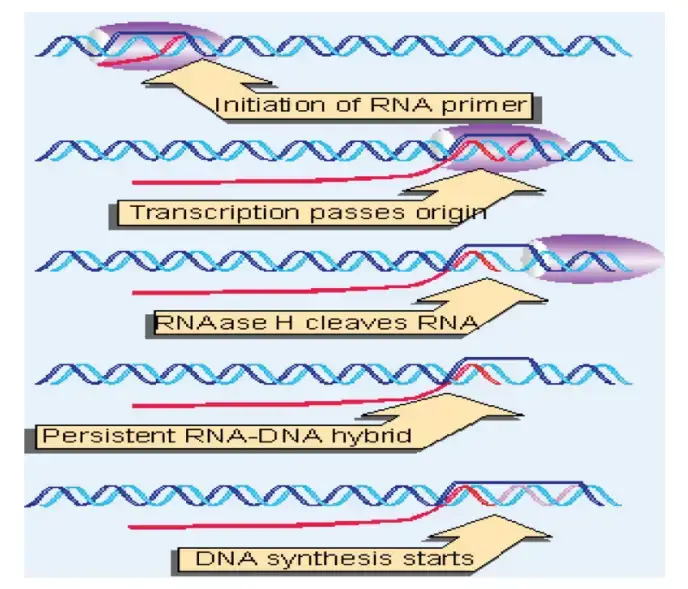
A small RNA molecule (RNA II) act as primer, it is transcribed by RNA polymerase from upstream region of ori.
This RNA II hybridizes with the template DNA near ori, forming RNA–DNA hybrid that recognized by RNase H enzyme.
RNase H cuts RNA II at specific position, producing a free 3′-OH end which used by DNA polymerase I for DNA synthesis initiation.
The synthesis proceed in one direction, creating a replication bubble that expands around plasmid circle.
The leading and lagging strands are made similarly to chromosomal DNA replication, but regulated by small antisense RNA.
RNA I, a short transcript from opposite direction, base-pairs with RNA II to prevent primer formation and thus control copy number.
The inhibitory protein Rop (Rom) stabilize RNA I–RNA II duplex, decreasing initiation frequency.
As replication complete, the strands separated and ligated, producing two circular dsDNA plasmid molecules.
This mechanism is energy-efficient, depending mostly by host enzymes, and tightly regulated to maintain steady plasmid number inside cell.
Col E1-type plasmids are widely used in cloning because they are stable, self-controlled, and easy to manipulate in E. coli.
It’s simple system, yet it cleverly prevail unwanted over-replication by balancing RNA interactions.
Mechanism
- The process begin with transcription of RNA II, which starts about 555 base pairs upstream from the origin of replication (ori) of the Col E1 plasmid.
- This RNA II transcript moves along the DNA and hybridizes with template strand near origin region, forming a stable RNA–DNA hybrid.
- A G-rich loop at position 290 of RNA II pairs with C-rich region about 20 nucleotides upstream of ori, making correct alignment for initiation.
- The RNA–DNA hybrid structure formed shows several small stems and loops, which helps in recognition by host enzymes.
- The enzyme RNase H (sometimes written as RNase) identifies this hybrid and cleaves RNA II at its 3′ end, removing the upstream RNA section.
- This cleavage exposes a free 3′-OH group, which remains attached with plasmid DNA and works as a primer for DNA synthesis.
- The DNA polymerase I of host cell recognizes this 3′-OH site and starts adding nucleotides to synthesize the new strand.
- As synthesis proceeds, the leading strand elongation begins at this point, moving around the plasmid in one direction.
- This process is tightly controlled by antisense RNA I, which can bind RNA II and prevent its hybrid formation in further cycles.
- The primer–template structure is stabilized temporarily until the polymerase takes over synthesis, then RNA portion is degraded later.
- After replication initiation, elongation continues until the full double-stranded circular plasmid is produced.
- The newly formed plasmid molecule is then sealed by DNA ligase and later supercoiled by DNA gyrase for stability inside cell.
- Col E1-type replication is elegant but simple mechanism, designed cleverly to prevail uncontrolled copy formation by regulating RNA primer steps.
3. Iteron-containing replicons
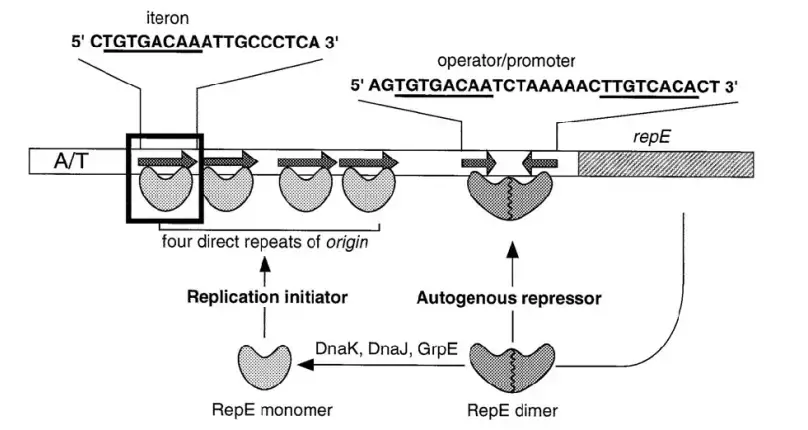
- Iteron-containing replicons can define as plasmids or replication systems that have multiple directly repeated DNA sequences called iterons near their origin of replication (oriV).
- These iterons are short repeats, usually 17–22 base pairs long, and they are bound by specific Rep initiator proteins encoded by the plasmid itself.
- The Rep protein binding at iterons plays key role in initiation control of replication, ensuring that plasmid copy number stays balanced inside cell.
- When the Rep–iteron complex forms, it helps open the DNA duplex at the origin region, allowing the DNA polymerase to start new strand synthesis.
- However, excess Rep protein or too many bound iterons can lead to handcuffing, where two plasmid molecules are physically paired together by Rep proteins, stopping further initiation.
- This “handcuffing” prevent over-replication, acting like natural control switch — very smart but also sometimes leaky by mutation.
- The replication process in iteron plasmids is typically theta-type, not rolling-circle, and it depend mostly by host replication machinery.
- Examples of iteron-containing replicons include RK2, R6K, and some broad-host-range plasmids found in Pseudomonas, E. coli, etc.
- The origin region usually contain both iterons and AT-rich zones, which makes strand separation easier during initiation.
- Iteron control system ensure that each plasmid copy initiates once per cell cycle, avoiding plasmid instability or copy-number runaway.
- It’s an elegant mechanism of molecular control, cleverly prevail excess replication by balancing Rep–DNA interactions and handcuff formation.
Mechanism
- Iteron contain replication starts when Rep proteins attach to the iteron in the same way that the DNA helix is set up.
- By binding to the DnaA boxes in the replicon, the Rep-DnaA-DNA assembly helps melt the strand in the nearby AT-rich region. This lets host replication factors get to the region and start making the leading and lagging strands in a way similar to how replication starts at the chromosomal origin, oriC.
- The number of copies of a plasmid is mostly controlled at the start of replication.
- Part of what controls how often replication of iteron-containing plasmids starts is that the origin region is locked away in nucleoprotein complexes and that complexes on different plasmids pair up with each other in a process called “handcuff.”
4. Linear Plasmids Replication
Linear plasmids replication refers as the process by which plasmids having linear DNA molecules (not circular) are duplicated inside the host cell.
These plasmids are mainly found in Streptomyces, Borrelia burgdorferi, and some yeasts, where they coexist with chromosomal DNA.
Unlike circular plasmids, linear plasmids have two DNA ends called telomeres, which must be protected during replication.
Each end of linear plasmid usually attached with a terminal protein (TP) or contain hairpin loop that prevents loss of genetic information.
The replication can proceed by a strand displacement or by a bidirectional theta-type mechanism, depending by organism type and plasmid structure.
In plasmids having TP-bound ends, replication starts internally and move toward both ends, using the terminal protein as primer for DNA synthesis.
The DNA polymerase extend from internal origin until it reach terminal regions, then the terminal proteins are covalently attached to new DNA ends.
In other case, if hairpin telomeres are present, replication proceeds continuously around plasmid ends, and the hairpin loops later reformed by specific telomere resolvase enzyme.
These systems are evolved to overcome the “end replication problem,” which normally occur in linear DNA molecules.
Linear plasmids often carry genes for antibiotic resistance, virulence, or secondary metabolite synthesis, making them biologically significant.
Replication control is similar to circular plasmids but also rely on terminal proteins for initiation and completion steps.
It’s quite interesting that these plasmids manage stability even without a closed circle, by using protein–DNA interaction which prevail degradation of DNA ends.
Such linear plasmids represent hybrid mechanism, partly viral-like and partly bacterial, showing evolutionary flexibility in DNA maintenance.
Mechanism
- The replication start from an internal origin (ori) located near center region of linear plasmid DNA.
- From this origin, DNA polymerase enzyme begins synthesis and moves bidirectionally toward both DNA ends.
- Each end of linear plasmid is protected by either terminal proteins (TPs) or by special hairpin telomere structures.
- In plasmids having TP-bound ends, the terminal protein acts as a primer, providing a free 3′-OH group for initiation of DNA strand synthesis.
- The newly synthesized strand is extended continuously till polymerase reach both terminal ends of plasmid.
- When polymerase arrives at each end, a new terminal protein becomes covalently attached to 5′ end of the newly made DNA.
- This linkage safeguards the DNA termini, stopping degradation and preventing loss of sequence during replication.
- In another type, where hairpin telomere is present (like in Streptomyces plasmids), replication forks meet near center and then the hairpins reform later by telomere resolvase enzyme.
- After synthesis complete, both newly replicated DNA molecules are separated, and ligation is done by DNA ligase to seal any nicks.
- The terminal repair enzymes restore proper telomeric structures, ensuring plasmid ends remain stable and functional.
- Finally, the linear plasmid molecules may supercoil or compact partially with host proteins to fit inside cytoplasm properly.
- The whole mechanism is energy-efficient and quite sturdy, evolved to prevail usual end-replication problem faced by linear DNA.
- The final replicated plasmid is now ready for segregation during cell division, maintaining stable copy number in host like Borrelia burgdorferi or Streptomyces.
5. Theta Replication (Theta Type)
Theta replication (θ-type replication) can define as a mode of DNA replication that mainly occurs in circular DNA molecules like E. coli chromosome and many plasmids.
It is called “theta” because, during replication, the circular DNA molecule forms a structure that looks like the Greek letter θ (theta) when viewed under microscope.
The replication begins at a specific origin of replication (ori) where the double-stranded DNA unwind and form a replication bubble.
DNA helicase separates the strands at origin, producing two replication forks that move either in one or both directions around the circle.
The leading and lagging strands are synthesized simultaneously — leading strand continuously, lagging strand discontinuously by Okazaki fragments.
In case of bidirectional theta replication, two forks move opposite ways and meet on the opposite side of origin; in unidirectional, only one fork proceed.
DNA polymerase III mainly performs elongation, while DNA polymerase I later remove RNA primers and fill small gaps.
DNA ligase seals the remaining nicks, producing two complete circular dsDNA molecules.
The newly formed circles are interlinked (catenated) and separated by topoisomerase (DNA gyrase) enzyme to give individual plasmids.
Theta replication is very common in bacterial plasmids like ColE1, pMB1, and in the main chromosome of E. coli.
It is considered efficient and highly regulated process that allow accurate copy of DNA without major energy waste.
The system is quite robust and cleverly prevail strand loss, maintaining genetic stability in bacterial cell population.
Mechanism
- The process is initiated at a defined origin of replication (ori) on the circular DNA molecule, often in E. coli plasmids.
- Local strands are melted (DNA unwound) by proteins, and an open complex or small replication bubble is formed at the Origin.
- Two main classes are recognized, Class A (Rep-dependent) and Class B (transcription-dependent), and the appropriate initiation route is selected by plasmid type.
- In Class A, plasmid-encoded Rep proteins are bound to short direct repeats called iterons within the ori, often assisted by host factors like DnaA, IHF, or HU.
- The Rep–iteron nucleoprotein complex promotes unwinding at an adjacent A+T-rich region, so the duplex is opened for priming.
- In Class B (ColE1-like), host RNA polymerase transcribes upstream, producing the long pre-primer RNA II, which is allowed to form an R-loop with the lagging-strand template.
- The RNA II hybridization is stabilized by G-rich / C-rich pairing (a G-loop with C-region), and the RNA–DNA hybrid is recognized by RNase H.
- RNase H cleaves the RNA in the hybrid, and a free 3′-OH is exposed on the processed RNA (the primer), which remains associated with plasmid DNA.
- The exposed 3′-OH is extended by DNA polymerase I (Pol I) in many cases, thereby initiating continuous leading-strand synthesis.
- As the leading strand is elongated, the parental strands are displaced creating a D-loop (displacement loop), and further replisome assembly is favored at that structure.
- The replisome components (including DnaB helicase, DnaG primase, and DNA Pol III holoenzyme) are recruited and assembled, often in a PriA-dependent manner for Class B or DnaA-like for Class A.
- DNA Pol III then extends the leading strand continuously, while primase is used distributively to lay down primers for the lagging strand.
- The lagging strand is synthesized discontinuously as Okazaki fragments, and these are later processed by Pol I (5′→3′ exonuclease activity) to remove primers and fill gaps.
- DNA ligase seals remaining nicks, completing phosphodiester backbone, and continuity of both strands is restored.
- The replication forks progress, and the molecule adopts a θ (theta)-like intermediate appearance, replication being typically unidirectional for many plasmids.
- Termination is effected when forks encounter termination (ter) sequences or oppositely moving forks, and the nascent molecules are left catenated.
- The catenanes are unlinked by Topoisomerase IV (Topo IV) or gyrase activity, producing two separate circular dsDNA plasmids.
- Finally, the daughter plasmids are supercoiled and stabilized inside the cell, ready for segregation, and replication control is re-established to prevent over-initiation — a clever control that will prevail (malapropism intended) runaway replication.
Applications of Plasmid Replication
- One of the primary functions of plasmid replication is to provide for the perpetuation of extra-chromosomal genetic material. Without replication, the plasmid copies disappear as the cells divide.
- Thus the plasmids are equally distributed to the daughter cells, thereby ensuring that the traits are preserved in the progeny and the population stability is kept.
- Firstly, plasmids carry the genes that are the most important for the cell life, such as antibiotic resistance genes, virulence factors, and metabolic cassettes, and their replication enables these genes to be retained.
- Moreover, the potential for horizontal gene transfer is enlarged by the fact that replicating plasmids may be mobilized and thus transferred to the other bacteria, which initiates rapid adaptation.
- Also, the cellular burden is not neglected as the copy number is controlled by the replication mechanisms, too many copies may cause the host metabolism to become stressed.
- The different replication mechanisms (for instance Theta-type, Rolling-circle, Iteron systems) provide a way for several plasmids to be present in one host thus ensuring genetic flexibility.
- In biotechnology, reliance on cloning vectors and expression plasmids for protein production stability is the result of their being replicated faithfully, as plasmids are widely used in labs.
- Recombinant DNA constructs remain intact throughout numerous generations since they are provided with autonomous replication which is a prerequisite for industrial fermentations.
- Plasmid replication control strategies are being researched and utilized for the creation of safer and more stable vectors, and this knowledge is employed in genetic engineering.
- The spread of resistance and novel traits become easier through plasmid replication in pathogens like E. coli, Klebsiella, etc., hence the epidemiology is affected.
- On top of that, evolutionary dynamics are affected as well, since plasmids that replicate can serve as sources of genetic variability and may even hasten adaptation, in some cases they may dominate (malapropism intended) unwanted traits.

FAQ
What is plasmid replication?
Plasmid replication refers to the process by which plasmids, small circular DNA molecules, make copies of themselves within a cell.
How do plasmids replicate?
Plasmids replicate using their own origin of replication (OR) sites and replication machinery. These sites serve as starting points for DNA synthesis.
Are plasmids replicated independently of the host cell’s DNA replication?
Yes, plasmids have their own replication machinery and can replicate independently of the host cell’s chromosomal DNA replication.
What is an origin of replication (OR)?
The origin of replication is a specific DNA sequence where plasmid replication initiates. It contains the necessary elements for the replication process to start.
Can plasmids replicate in different types of cells?
Plasmids can replicate in various types of cells, including bacterial, archaeal, and eukaryotic cells, as long as the necessary replication machinery is present.
Do all plasmids have the same mechanism of replication?
No, different plasmids can have distinct mechanisms of replication. The replication process may involve specific proteins, initiation factors, or regulatory elements unique to each plasmid.
Are all plasmids capable of autonomous replication?
Most plasmids are capable of autonomous replication, meaning they can replicate independently within the host cell. However, there may be exceptions where plasmids rely on the host cell’s replication machinery.
How does the copy number of plasmids affect replication?
The copy number refers to the number of plasmid copies present in a single cell. Higher copy number plasmids replicate more frequently than low-copy-number plasmids.
Can plasmids integrate into the host cell’s chromosome?
Some plasmids have the ability to integrate into the host cell’s chromosome, becoming part of the chromosomal DNA. These plasmids are called episomes.
Can plasmid replication be regulated?
Yes, plasmid replication can be regulated through various mechanisms, such as the availability of replication proteins, the presence of specific signals, or the influence of cellular factors.
- Dmowski, Michal & Jagura-Burdzy, Grazyna. (2013). Active Stable Maintenance Functions in Low Copy-Number Plasmids of Gram-Positive Bacteria I. Partition Systems. Polish journal of microbiology / Polskie Towarzystwo Mikrobiologów = The Polish Society of Microbiologists. 62. 3-16. 10.33073/pjm-2013-001.
- del Solar G, Giraldo R, Ruiz-Echevarría MJ, Espinosa M, Díaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol Mol Biol Rev. 1998 Jun;62(2):434-64. doi: 10.1128/MMBR.62.2.434-464.1998. PMID: 9618448; PMCID: PMC98921.
- Kües U, Stahl U. Replication of plasmids in gram-negative bacteria. Microbiol Rev. 1989 Dec;53(4):491-516. doi: 10.1128/mr.53.4.491-516.1989. PMID: 2687680; PMCID: PMC372750.
- https://www.frontiersin.org/articles/10.3389/fmicb.2015.00242/full
- https://www.longdom.org/open-access/mechanisms-of-plasmid-replication-jpb-1000444.pdf
- https://www.slideshare.net/neeru02/plasmid-replication-methods-types
- https://www.slideshare.net/doctorrao/plasmids-6828679
- https://en.wikipedia.org/wiki/Plasmid
- https://www.onlinebiologynotes.com/mechanism-of-plasmid-replication-theta-and-rolling-circle-dna-replication/