What is Operon?
- An operon represents a functional unit of DNA that comprises a cluster of genes under the control of a singular promoter. This genetic configuration is predominantly found in prokaryotes, including bacteria and archaea, but is not a feature of eukaryotic organisms. The genes within an operon are transcribed collectively into a single mRNA strand. This mRNA can then undergo translation in the cytoplasm or experience splicing to produce monocistronic mRNAs, which are then individually translated. Consequently, the genes within an operon are either co-expressed or not expressed at all.
- The concept of the operon was introduced in 1961 by François Jacob and Jacques Monod, marking a pivotal moment in molecular biology. The primary operon to be elucidated was the lac operon in E. coli. For their groundbreaking work on the operon and virus synthesis, Jacob, André Michel Lwoff, and Monod were awarded the Nobel Prize in Physiology and Medicine in 1965.
- Operons are not exclusive to prokaryotes. Evidence from the early 1990s suggests the presence of operons in certain eukaryotes, including nematodes like C. elegans and the fruit fly, Drosophila melanogaster. Additionally, viruses, such as bacteriophages, also exhibit operons. For instance, T7 phages possess two operons, with the first coding for various products, including a unique T7 RNA polymerase. This polymerase can then transcribe the second operon, which contains a lysis gene designed to rupture the host cell.
- The structural genes within an operon are organized under a unified promoter and are regulated by a shared operator. This configuration allows for the genes to be transcribed into a singular mRNA molecule. The regulatory elements of an operon, which can include repressors, corepressors, and activators, might not be encoded by the operon itself. The positioning and state of these regulatory elements, in conjunction with the promoter, operator, and structural DNA sequences, can influence the outcomes of specific mutations.
- Furthermore, operons are associated with other regulatory groups such as regulons, stimulons, and modulons. While an operon consists of genes regulated by a shared operator, regulons encompass genes regulated by a singular regulatory protein. Stimulons, on the other hand, comprise genes regulated by a specific cellular stimulus. The term “operon” is believed to have been derived from the action verb “to operate.”
- In summary, operons play a crucial role in the coordinated regulation of gene expression, particularly in prokaryotic organisms. Their discovery and understanding have significantly advanced our knowledge of molecular biology and genetic regulation.
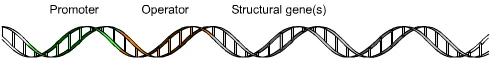
Definition of Operon
An operon is a functional unit of DNA in prokaryotes that contains a cluster of genes under the control of a single promoter, allowing for coordinated expression of related genes.
General structure of Operon
The operon is a fundamental genetic regulatory system found in prokaryotes, consisting of several key DNA components that work in tandem to control gene expression. Its general structure comprises:
- Promoter: This is a specific nucleotide sequence that acts as a starting point for gene transcription. The promoter is recognized by the RNA polymerase enzyme, which initiates the process of transcription. By determining which genes undergo transcription into messenger RNA (mRNA), the promoter indirectly dictates the proteins synthesized by the cell.
- Operator: Positioned typically adjacent to the promoter, the operator is a DNA segment that can bind a repressor protein. This binding can physically hinder the RNA polymerase from transcribing the genes. In certain operons, such as the lac operon, there might be multiple operators. The presence of repressors or corepressors can influence the operon’s activity. For instance, a corepressor can enhance the repressor’s ability to bind to the operator, while an inducer can displace the repressor, leading to an active operon.
- Structural Genes: These are the primary genes within the operon that are co-regulated. They encode the primary sequences of proteins that participate in specific metabolic pathways or cellular functions.
In addition to these core components, an operon’s function can be influenced by a Regulatory Gene. Although not always located within the operon itself, this gene is continuously expressed and produces repressor proteins. The position of the regulatory gene is not fixed; it can be distant from the operon it regulates.
Furthermore, external molecules, such as inducers and corepressors, play pivotal roles in modulating operon activity. An inducer can remove a repressor from the operator, activating the operon. Conversely, a corepressor can facilitate the repressor’s binding to the operator, leading to transcriptional repression.
In essence, an operon is a sophisticated genetic module comprising structural genes and regulatory elements, including the promoter and operator. These components collaboratively determine the transcriptional state of the operon, allowing the cell to respond dynamically to environmental cues or internal signals.
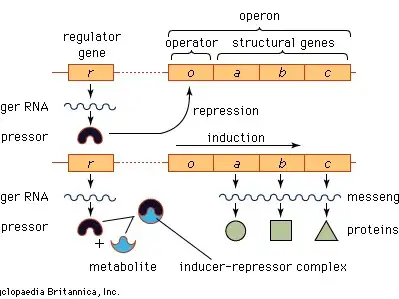
Regulation of Operon
The regulation of an operon is a sophisticated mechanism that allows organisms to modulate gene expression in response to environmental fluctuations. This gene regulation ensures that specific genes are expressed only when necessary, optimizing energy consumption and cellular function. The regulation of operons can be categorized into negative or positive control, each further classified by induction or repression.
- Negative Control:
- Negative Inducible Operons: In this system, a repressor protein, produced by a regulator gene, is typically bound to the operator, inhibiting the transcription of genes within the operon. When an inducer molecule is present, it interacts with the repressor, altering its shape and preventing it from binding to the operator. This de-repression allows the genes to be transcribed. The lac operon, regulated by the inducer allolactose, exemplifies a negatively controlled inducible operon.
- Negative Repressible Operons: Here, the operon is usually active, with genes being transcribed. The repressor protein, in its default state, cannot bind to the operator. However, when a specific molecule, termed a corepressor, interacts with the repressor, it induces a conformational change, enabling the repressor to bind to the operator and halt transcription. The trp operon, responsible for tryptophan synthesis, operates as a negatively controlled repressible operon, with tryptophan acting as the corepressor.
- Positive Control:
- Positive Inducible Operons: In this configuration, an activator protein is inherently unable to bind to the relevant DNA segment. Upon interaction with an inducer, the activator undergoes a structural change, enabling it to bind to the DNA and stimulate transcription. The MerR family of transcriptional activators serves as an example of positive inducible operons.
- Positive Repressible Operons: Typically, activator proteins are bound to the relevant DNA segment, promoting transcription. However, when these activators bind to an inhibitor molecule, they are rendered incapable of DNA binding, leading to transcriptional repression.
Types of Operon
There are present two different types of operon such as;
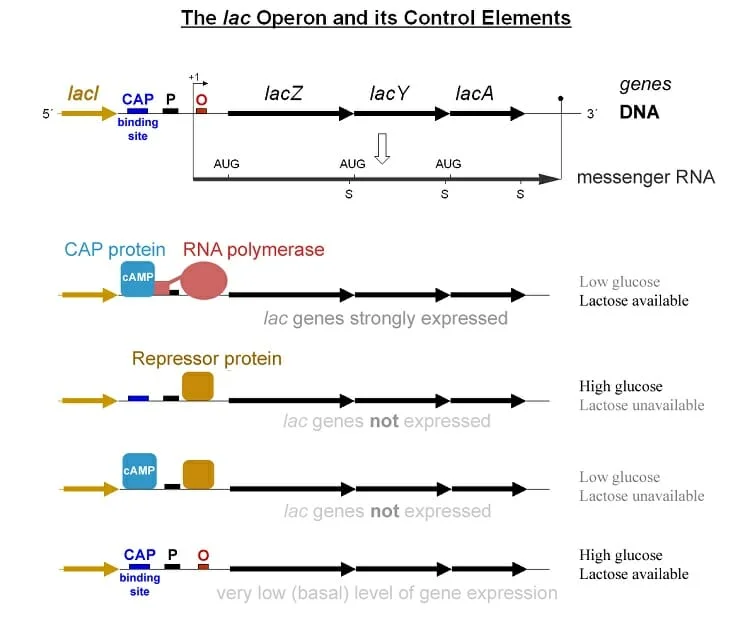
1. Lac Operon
The lac operon, predominantly found in E. coli and certain other bacteria, is a gene set specifically designed for the uptake and metabolism of lactose. This operon comprises three primary structural genes:
- lacZ: Encodes for β-galactosidase, an enzyme responsible for cleaving lactose into its constituent sugars, galactose and glucose.
- lacY: Produces lac permease, a transmembrane protein essential for the uptake of lactose into the cell.
- lacA: Encodes for a transacetylase that transfers an acetyl group from coenzyme A (CoA) to galactosides.
Adjacent to these structural genes, at the 5′ end relative to lacZ, is the lacI gene. This gene produces the lac operon’s repressor, which functions independently of the structural genes. The promoter, located ahead of the lacZ gene, sees its expression modulated by both the LacI repressor and the catabolite activator protein (CAP), also referred to as the cAMP receptor protein (CRP).
The lac operon’s expression is intricately regulated based on external nutrient levels. Specifically, it is activated when external lactose concentrations are high and glucose concentrations are low. This is because E. coli preferentially metabolizes glucose. Thus, the genes responsible for lactose metabolism are only activated when lactose is abundant and glucose is scarce.
The lac operon serves as a quintessential example of an inducible operon. In the absence of lactose, the operator is inhibited by a repressor protein. This operon consists of a promoter with an operator and the three aforementioned genes (lacZ, lacY, and lacA) that play roles in lactose breakdown. The regulatory gene, lacI, situated close to the promoter region, governs the lac operon’s expression. It encodes an allosteric repressor protein that typically keeps the lac operon inactive.
For the lac operon to be activated, an inducer molecule, allolactose (an isomer of lactose), must neutralize the repressor protein. When lactose and its isomer are present, allolactose binds to the repressor protein’s allosteric sites, altering its shape and deactivating it. This allows RNA polymerase to bind to the promoter, initiating transcription and facilitating the synthesis of the lactose degradation genes.
Furthermore, the lac operon is subject to positive gene regulation. While repressor protein removal is essential for the synthesis of lacZ, lacY, and lacA genes, the gene expression remains relatively low. The gene expression level is influenced by the cell’s glucose concentration, regulated by the catabolite activator protein (CAP). When glucose levels are low, cyclic AMP concentrations rise, activating CAP. This activated CAP binds to the lac operon promoter, enhancing gene expression. Conversely, as glucose levels rise, cyclic AMP concentrations drop, leading to CAP’s detachment from the promoter. This results in reduced transcription or complete cessation if the repressor protein rebinds.
lac Regulation
The regulation of the lac operon is a sophisticated process that involves both negative and positive control mechanisms to ensure the efficient utilization of energy sources by the cell.
- Negative Control via the LacI Repressor: The LacI repressor is continuously synthesized from its distinct promoter. Upon formation, it assembles into a homotetramer, which binds to the DNA, specifically inhibiting the RNA polymerase from transcribing the three structural genes of the lac operon. This repression exemplifies negative control.
- Positive Regulation via cAMP–CRP Complex: When glucose levels outside the cell are low, there is a surge in the intracellular production of cyclic AMP (cAMP). This cAMP binds to the catabolite activator protein (CRP), forming the cAMP–CRP complex. This complex then attaches to a specific DNA site located upstream of the lac promoter. By doing so, it amplifies the binding and stabilizes the RNA polymerase-DNA open complex at the onset of transcription. This mechanism, termed catabolite repression, represents positive regulation.
- Lactose Uptake and Allolactose Formation: In conditions where glucose is scarce but lactose is abundant, the LacY permease facilitates the transport of external lactose into the cell. Once inside, the basal levels of β-galactosidase convert lactose to allolactose. This allolactose then interacts with each LacI monomer, inducing a structural change that lifts the repression from the lac operon. As a result, transcription and subsequent translation are initiated.
- Glucose’s Negative Impact on Lactose Uptake: If the external environment is rich in glucose, it interacts with the LacY permease, reducing the uptake of lactose into the cell. This interaction hinders the formation of allolactose, thereby negatively affecting the induction of the lac operon genes.
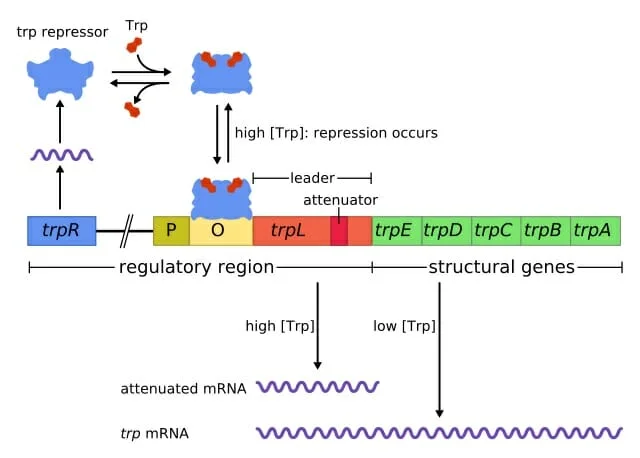
2. Trp Operon
The Trp operon, a crucial regulatory system in bacteria, orchestrates the synthesis of the amino acid tryptophan based on its environmental availability. Located in prokaryotes, this operon comprises five genes encoding enzymes essential for tryptophan biosynthesis. These genes are trpE, trpD, trpC, trpA, and trpB, which collectively facilitate the transformation of chorismic acid into tryptophan. Additionally, the trpL gene encodes a short oligopeptide involved in attenuation. The operon’s regulation is initiated at its promoter, followed by the attenuator, situated upstream of the trpE gene.
Two primary mechanisms regulate the Trp operon:
- Repressor-mediated Control: The trpR gene, located distally from the Trp operon, encodes the TrpR repressor protein. This protein, although constantly present, is synthesized in an inactive state. When tryptophan levels within the cell rise, it acts as a corepressor, binding to the TrpR repressor. This binding induces a conformational change in the TrpR protein, enabling it to bind to the operon’s operator. Consequently, this binding obstructs the RNA polymerase’s access to the promoter, inhibiting transcription. This mechanism exemplifies a repressible operon, where the operon remains active unless deactivated by a specific condition, in this case, elevated tryptophan levels.
- Attenuation: This secondary regulatory mechanism can further suppress the operon’s expression. While the TrpR repressor can reduce transcription by a factor of 70, attenuation can amplify this reduction tenfold, resulting in a cumulative 700-fold decrease in expression. This intricate process is feasible in prokaryotes due to the simultaneous occurrence of transcription and translation. The leader transcript, trpL, at the operon’s commencement, contains sequences that can form various stem-loop structures. The presence of the 3-4 stem-loop, in conjunction with the attenuator, acts as a transcriptional terminator. An intriguing aspect of this mechanism is the leader peptide encoded by part of the trpL transcript. This peptide, enriched with tryptophan residues, serves as a sensor for intracellular tryptophan levels. Low tryptophan levels cause ribosomal stalling during this peptide’s translation, leading to the formation of the 2-3 stem-loop, which prevents transcription termination. Conversely, abundant tryptophan ensures uninterrupted translation of the leader peptide, resulting in the formation of the 3-4 termination structure, thereby halting transcription.
Operon Functions
Operons are fundamental genetic regulatory systems found primarily in prokaryotes, such as bacteria. They play a crucial role in the coordinated regulation of gene expression. Here are the primary functions of operons:
- Coordinated Gene Regulation: Operons allow for the simultaneous regulation of a group of functionally related genes. When a specific stimulus is present, all the genes within an operon can be transcribed together, ensuring a coordinated response.
- Efficient Resource Utilization: By grouping genes that have a common function or are part of the same metabolic pathway, the cell can efficiently turn on or off entire pathways in response to environmental changes. This ensures that energy and resources are not wasted on producing unnecessary proteins.
- Simplification of Gene Regulation: Instead of requiring individual regulatory mechanisms for each gene, operons allow for a single regulatory mechanism to control the expression of multiple genes. This simplifies the regulatory process and conserves genetic space.
- Response to Environmental Changes: Operons enable bacteria to quickly adapt to changes in their environment. For example, if a nutrient becomes available, the genes required to metabolize that nutrient can be rapidly turned on.
- Negative and Positive Regulation:
- Negative Control: Operons can be regulated by repressors that inhibit gene transcription. This can be further classified into repressible and inducible operons.
- Positive Control: Some operons are activated by specific activator proteins that enhance gene transcription.
- Flexibility in Gene Expression: Depending on the cellular needs and environmental conditions, operons can be fully active, partially active, or completely inactive. This flexibility allows for fine-tuned regulation of gene expression.
- Protection Against Random Genetic Insertions: By clustering essential genes together, operons may reduce the likelihood of harmful genetic insertions that could disrupt important cellular functions.
In essence, operons provide a streamlined and efficient mechanism for gene regulation, allowing prokaryotic organisms to swiftly adapt to their ever-changing environments.
Quiz
What is an operon?
a) A type of virus
b) A sequence of DNA that codes for a protein
c) A set of adjacent genes that are regulated together
d) A type of enzyme
Which of the following is NOT a component of an operon?
a) Promoter
b) Operator
c) Repressor
d) Ribosome
The Lac operon is primarily involved in the metabolism of:
a) Glucose
b) Tryptophan
c) Lactose
d) Histidine
Which type of operon is usually “on” but can be turned “off” by a specific molecule?
a) Repressible operon
b) Inducible operon
c) Positive operon
d) Negative operon
In the absence of lactose, the lac operon is:
a) Active and producing enzymes
b) Inactive due to the binding of a repressor protein
c) Active due to the binding of an activator protein
d) Inactive due to the absence of an activator protein
The Trp operon is responsible for the synthesis of which amino acid?
a) Lysine
b) Methionine
c) Tryptophan
d) Tyrosine
Which molecule acts as an inducer in the lac operon?
a) Glucose
b) Tryptophan
c) Allolactose
d) Histidine
In which organism was the concept of the operon first described?
a) Saccharomyces cerevisiae
b) Escherichia coli
c) Homo sapiens
d) Drosophila melanogaster
Positive control in operon regulation means:
a) The operon is turned off by a repressor
b) The operon is turned on by an activator
c) The operon is turned off by an activator
d) The operon is turned on by a repressor
Which of the following operons is an example of a repressible operon?
a) Lac operon
b) Trp operon
c) His operon
d) Gal operon
FAQ
What is an operon?
An operon is a functional unit of DNA containing multiple genes, which are regulated together by a single promoter and operator.
Why are operons important in bacteria?
Operons allow bacteria to efficiently regulate the expression of genes in response to environmental changes, ensuring that only necessary genes are active at any given time.
What is the difference between inducible and repressible operons?
Inducible operons are usually off and can be turned on by a specific substance, while repressible operons are typically on and can be turned off by a specific substance.
How does the lac operon work?
The lac operon is an inducible operon that is activated in the presence of lactose. When lactose is present, it inhibits a repressor protein from binding to the operon, allowing the genes to be transcribed.
What role does the repressor protein play in operon regulation?
The repressor protein can bind to the operator region of an operon, preventing RNA polymerase from transcribing the genes. The binding of the repressor can be influenced by specific molecules, allowing for gene regulation.
Why is the trp operon considered a repressible operon?
The trp operon is involved in the synthesis of tryptophan. When tryptophan levels are high in the cell, it acts as a co-repressor, enabling the repressor protein to bind to the operon and turn it off.
What is catabolite repression?
Catabolite repression is a form of regulation where the presence of a preferred nutrient (like glucose) represses the expression of genes involved in the metabolism of alternative nutrients.
How do positive and negative control differ in operon regulation?
Positive control involves the activation of gene expression by a regulatory protein, while negative control involves the repression of gene expression by a regulatory protein.
Are operons found in eukaryotes?
Operons are primarily a feature of prokaryotes. While there are a few examples of operon-like structures in eukaryotes, they are not as common or well-defined as in prokaryotes.
How does attenuation work in the trp operon?
Attenuation is a regulatory mechanism in the trp operon where the rate of transcription is influenced by the rate of translation. Depending on tryptophan levels, different stem-loop structures form in the mRNA, which can either promote or terminate transcription.
References
- Ramos, J. L., García-Salamanca, A., Molina-Santiago, C., & Udaondo, Z. (2013). Operon. Brenner’s Encyclopedia of Genetics, 176–180. doi:10.1016/b978-0-12-374984-0.01096-2
- Picknett, T. M., & Brenner, S. (2001). Operon. Encyclopedia of Genetics, 1377. doi:10.1006/rwgn.2001.0936
- Osbourn AE, Field B. Operons. Cell Mol Life Sci. 2009 Dec;66(23):3755-75. doi: 10.1007/s00018-009-0114-3. Epub 2009 Aug 7. PMID: 19662496; PMCID: PMC2776167.
- https://biologydictionary.net/operon/
- https://brianmccauley.net/bio-6b/6b-lecture/operons
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.