Okazaki fragment is the short segment of DNA that is synthesized on the lagging strand during DNA replication. It is formed because the DNA polymerase can extend the new strand only in the 5’–3’ direction, so the strand that is oriented in the 3’–5’ direction is produced in a discontinuous manner.
It is initiated by an RNA primer at the 5’ end, and the fragment is then extended by DNA polymerase. These are later joined to form a continuous daughter strand by the enzyme DNA ligase. It is the process that explains how the lagging strand is able to grow even though its orientation is opposite to the movement of the replication fork.
These fragments were first observed while studying the replication of bacteriophage DNA in Escherichia coli. It was during the late 1960s that Reiji Okazaki and Tsuneko Okazaki identified that newly synthesized DNA on the lagging strand is produced as small pieces before being combined. In prokaryotes, the fragments are longer and usually range from 1000 to 2000 nucleotides, while in eukaryotes the size is much shorter, about 150 to 200 nucleotides. It is the size variation that reflects the differences in replication machinery between the two systems.
The formation of Okazaki fragments is explained through a discontinuous mode of replication. In this step the daughter strand that is oriented in the 3’–5’ direction is synthesized as repeated short fragments in the opposite 5’–3’ direction, and after synthesis they are linked together to form the complete strand. This is referred to as semidiscontinuous replication.
The explanation was supported by pulse-chase experiments where short radioactive DNA was first detected as small fragments and later appeared in larger DNA after the fragments became joined. It proved that the lagging strand is not formed as one continuous piece but rather originates through multiple cycles of priming, extension, and ligation.
Okazaki Fragments Definition
Okazaki fragments are short DNA sequences synthesized discontinuously on the lagging strand during DNA replication, which are later joined together to form a continuous strand.
Why do okazaki fragments form?
Okazaki fragments are formed because the DNA polymerase enzyme is restricted to synthesize the new DNA strand only in the 5’–3’ direction, and this creates a difficulty when both strands of the double helix are copied. It is the nature of DNA that the two strands run in antiparallel directions, so when the replication fork opens, one template strand is oriented in the 3’–5’ direction and is copied continuously.
This is referred to as the leading strand. The other template strand runs in the 5’–3’ direction, and because the polymerase cannot extend in the reverse direction, a continuous synthesis is not possible on this strand.
To solve this problem, the cell synthesizes the lagging strand in a discontinuous manner. In this step short stretches of DNA are produced each time the helicase exposes a new segment of the template. The synthesis always begins from a new RNA primer, and the DNA polymerase extends it in the usual 5’–3’ direction but away from the opening fork. These short pieces are the Okazaki fragments. It is the process that allows the lagging strand to grow even though the fork moves in the opposite direction.
These fragments form because the replication machinery must keep the same direction of polymerase activity on both strands. Since the lagging template cannot be copied in one stretch, the discontinuous synthesis becomes the only way to maintain replication on both sides of the fork.
After formation, the fragments are joined to form a complete daughter strand, which explains how the antiparallel structure of DNA is accommodated during replication.
Why Okazaki fragments are discontinuous?
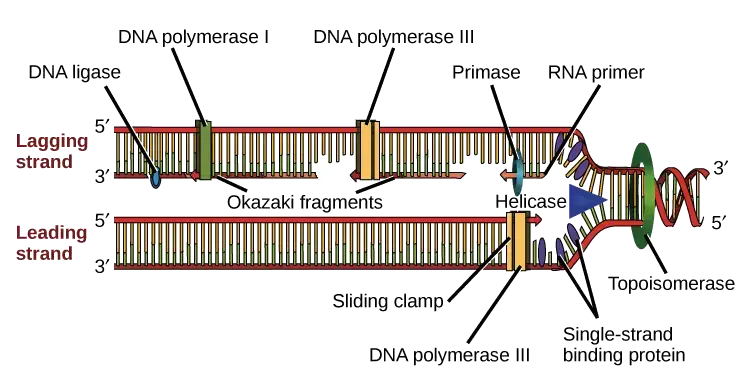
Okazaki fragments are discontinuous because the two strands of DNA run in antiparallel direction, and it is the major reason that only one strand is synthesized in a continuous way. DNA polymerase can extend the new DNA strand only in 5′→3′ direction, so when the replication fork opens, the leading strand is formed smoothly as the fork moves forward. It is the 3′→5′ template which allow the polymerase to move continuously along the fork.
The lagging strand template runs in the 5′→3′ direction, so the polymerase cannot move in the same direction as fork movement. This process occurs when the polymerase is forced to synthesize the new strand in short stretches away from the fork. In this step primase forms repeated RNA primers, and small stretches of DNA are extended from each primer. These are called Okazaki fragments, and two or more fragments is always separated by short gaps.
As the fork keeps on unwinding, the polymerase is again required to shift near the fork region and start a new fragment. It is the reason the synthesis on the lagging strand is fragmented and not continuous. After formation, these fragments are later joined by DNA ligase to form the complete daughter strand.
Formation of Okazaki Fragments – How are okazaki fragments synthesized?
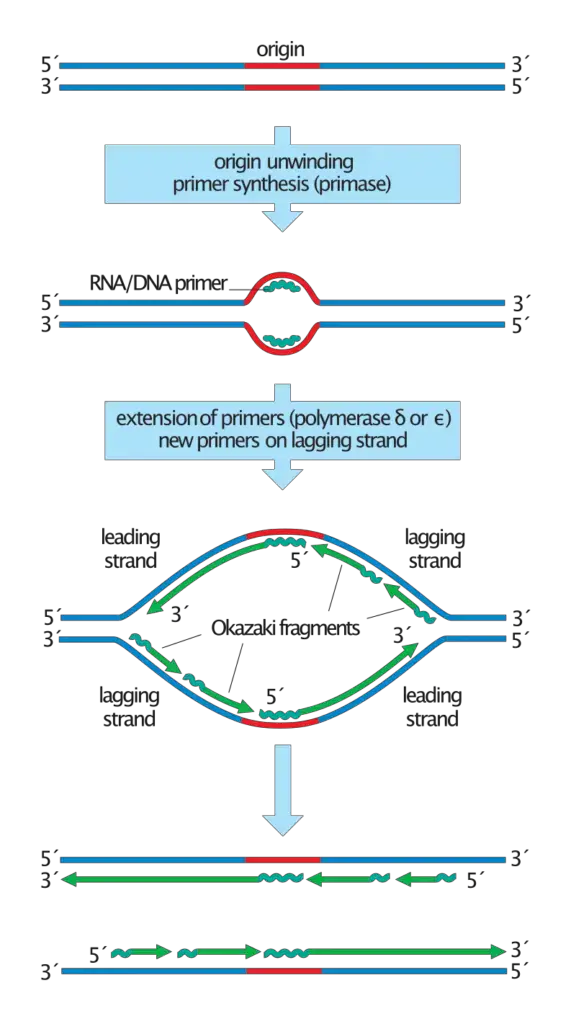
This occurs because DNA polymerase can add nucleotides only in the 5′→3′ direction, while the lagging strand template is oriented in the opposite way. Due to this, the synthesis cannot proceed continuously on this strand.
The replication fork is first created when the parental DNA is unwound. The single-stranded region of lagging strand is exposed and it is stabilized by binding proteins which prevent folding or reannealing. This exposed template acts as the surface on which short fragments will be formed.
A small RNA primer is prepared on the lagging strand. It is synthesized by primase enzyme and it gives the free 3′-OH group required for DNA chain initiation. In eukaryotic cells this primase is associated with DNA polymerase α and after preparing the RNA segment it also adds a short stretch of DNA. This mixed primer is essential because each new fragment starts only after a primer is made.
Once the primer is present, a change in polymerase occurs. The initiating enzyme is removed and a sliding clamp is attached around the primer region. This clamp helps in holding the elongating polymerase for stable synthesis. The clamp loader proteins assist in placing the clamp at the correct site.
DNA polymerase δ then starts synthesis from the 3′-end of the primer. It always moves in 5′→3′ direction but since the DNA fork moves in the opposite direction, the new fragment is formed away from the fork. The fragments are short in size, usually around 100–200 nucleotides in eukaryotes. These fragments together form the discontinuous pattern of the lagging strand.
As the polymerase continues, it reaches the starting point of the previous fragment. At this point the enzyme does not stop immediately but displaces the small RNA-DNA piece which lies ahead. This displaced region becomes a short flap structure. The formation of flap slows down the polymerase activity and marks the end of the fragment.
The flap is then removed by specific endonuclease enzymes and the remaining gap is filled by DNA. Later, the adjacent fragments are joined by DNA ligase forming a single continuous DNA strand. This whole sequence of events is referred to as formation of Okazaki fragments and it is essential for maintaining the proper replication of the lagging strand.
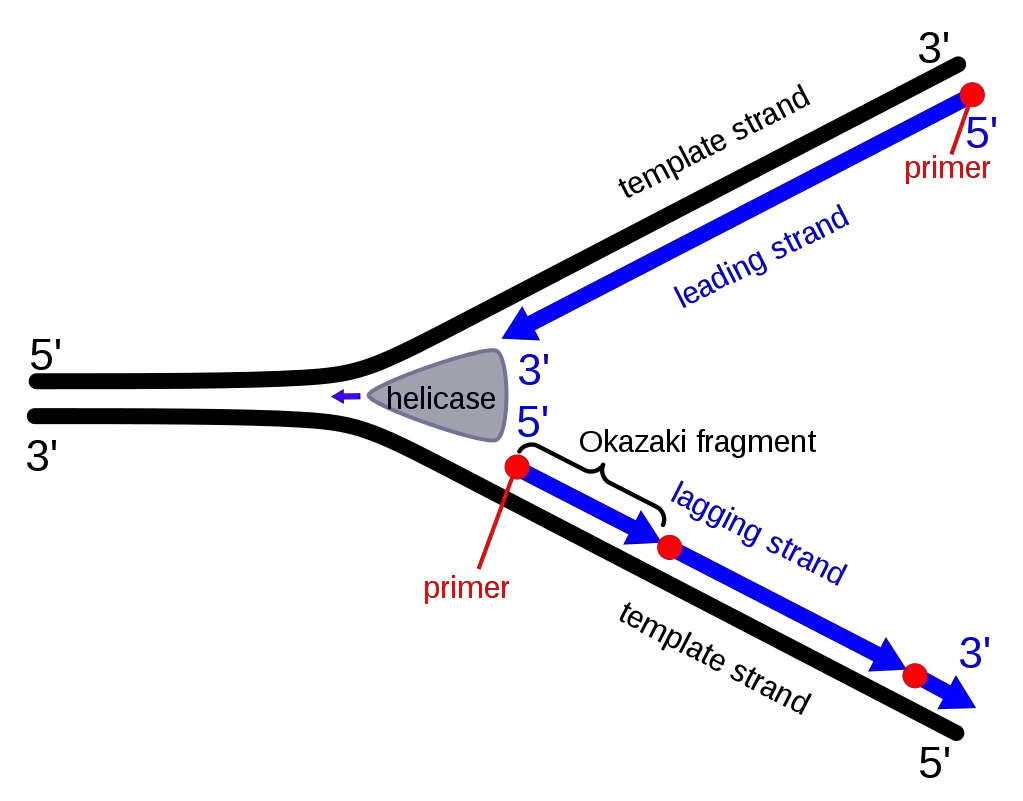
Formation of Okazaki Fragments: A Step-by-Step Process
- The replication fork is formed when the DNA double helix is unwound by helicase. The exposed single strand is stabilized by binding proteins which prevent folding of the strand.
- A short region of the single strand is cleared for primase activity. It is the step where primase enzyme gets access to the template for starting the new fragment.
- Primase synthesizes a short RNA primer. This primer provides the free 3′-OH end which is required for beginning the synthesis of the fragment.
- A sliding clamp is loaded around the primer region. The clamp loader complex helps in attaching the clamp at this point. The clamp is used to hold the polymerase for further elongation.
- DNA polymerase now extends the primer in the 5′→3′ direction. The synthesis always moves away from the previous fragment and goes towards the replication fork.
- As the fork moves ahead, new primers are added again near the fork. This make the synthesis on lagging strand discontinuous and formed in short units.
- When the polymerase reaches the 5′ end of the earlier fragment, it is released and the clamp is removed. A new clamp is placed at the next primer and the polymerase starts the next fragment. This cycle continues in a repeated manner.
- The RNA primers are removed later by specific enzymes. In this step, endonucleases cut the primer region and produce a small gap between the fragments.
- The gap is filled by DNA polymerase. It adds the required nucleotides and completes the DNA portion in place of the primer.
- DNA ligase seals the nick that remain between the two fragments. This form a continuous strand on the lagging side.
- The process repeats many times along the template. These short fragments together complete the semi-discontinuous synthesis of the lagging strand.
Significance of Okazaki fragment
- It is important for solving the problem created by the antiparallel nature of DNA. The fragments allow the replication machinery to synthesize the lagging strand in the required 5′→3′ direction.
- These fragments make the replication process semi-discontinuous. One strand is made in a continuous way, while the lagging strand is produced in short units.
- The fragments act as the main building blocks of the lagging strand. Each small segment is later joined to form the complete daughter strand.
- The joining of the fragments by DNA ligase is essential for forming a continuous DNA molecule. Without this, the strand remain broken.
- The repetitive initiation of each fragment makes the replication process more complex. It is controlled by several enzymes which manage the maturation steps.
- Millions of such fragments are formed in each cell cycle, especially in higher organisms. Their proper maturation is required for normal cell division and growth.
- The maturation involves removal of RNA primer and replacing it with DNA. Enzymes like polymerase, flap endonuclease and ligase are used in this step.
- This process also helps in correcting the initial errors introduced by the primase-polymerase complex. It increases the accuracy of the lagging strand.
- Proper formation and processing of the fragments is necessary for maintaining the genomic stability. Failure in this step can produce unsealed nicks and breaks.
- Defects in fragment maturation can cause chromosome damage. This may result in genetic disorders and sometimes lead to cancer development.
- The enzymes used in fragment processing are also used by DNA repair pathways. Thus the fragments act as intermediates which connect replication with repair mechanisms.
- FEN1, which removes flap structures during fragment maturation, also acts in long-patch repair systems. This shows the shared mechanism for maintaining DNA integrity.
Enzymes involved in Okazaki fragments formation
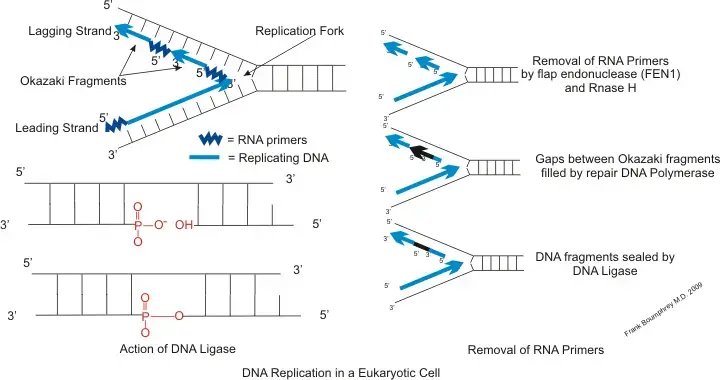
- Primase enzyme is used for synthesizing the short RNA primer on the lagging strand. It provides the 3′-OH group that is required to start the fragment.
- DNA polymerase α complex is responsible for extending this RNA primer by adding a short stretch of DNA. It forms the RNA-DNA initiator part which starts each Okazaki fragment.
- Replication Factor C (RFC) acts as the clamp loader. It recognizes the primer-template junction and helps in removing polymerase α from the site.
- PCNA works as the sliding clamp. It is attached around the DNA and holds the elongation polymerase for stable synthesis of the fragment.
- DNA polymerase δ is the main enzyme which forms the bulk of the Okazaki fragment. It extends the primer in 5′→3′ direction and also performs strand displacement when it meets the previous fragment.
- In prokaryotes, DNA polymerase III holoenzyme does the elongation step. It synthesizes the lagging strand fragments till it reaches the earlier primer.
- RNase H removes most part of the RNA primer. It cuts the RNA that remains attached to the DNA strand.
- Flap endonuclease 1 (FEN1) cleaves the small flap that is produced by strand displacement. This removes the remaining primer and creates the nick required for the next step.
- DNA polymerase I is used in prokaryotes for primer removal and gap filling. It removes the RNA using its exonuclease activity and adds DNA in the same process.
- DNA ligase I seals the final nick between two fragments. It joins the DNA pieces and forms the continuous lagging strand.
- Dna2 enzyme acts mainly when long flap structures are formed. It cuts these long flaps so that FEN1 can finish the processing.
- Pif1 helicase can make the displacement activity stronger. It helps in creating long flaps in yeast cells, requiring Dna2 for further action.
- Replication Protein A (RPA) binds to single-stranded DNA. It stabilizes the template and also controls the long flap pathway during fragment maturation.
- Exonuclease 1 (Exo1) works as a backup enzyme. It can remove flap structures if FEN1 activity becomes insufficient.
Which enzyme joins okazaki fragments?
The enzyme responsible for joining the Okazaki fragments is DNA ligase. In eukaryotic cells it is mostly done by DNA ligase I. It is the enzyme that seals the nick which remain after primer removal and DNA filling.
Mechanism
1. Preparation of the ligatable nick
It is necessary that the RNA primer at the 5′ end is removed completely and then the gap is filled with DNA. This is carried out by DNA polymerase δ and FEN1 in eukaryotes. In this step the polymerase displaces the primer forming a short flap and the flap is cleaved by FEN1. The activity of these enzyme is repeated until two DNA–DNA ends is produced. These are abutted ends leaving only a single nick in the phosphodiester backbone.
2. Binding of DNA ligase I
The nick contain a 3′–OH group at the downstream fragment and a 5′–phosphate at the upstream fragment. DNA ligase I is recruited to this site. It is the enzyme which recognize this nicked structure. It binds at the junction so that the phosphodiester bond can be formed.
3. Formation of the phosphodiester bond
It is the process where ligase catalyze the joining reaction. The enzyme activates the 5′–phosphate end by forming a ligase–AMP complex, and then the AMP is transferred to the DNA. After this activation the 3′–OH attack the activated 5′–P forming the phosphodiester linkage. The DNA becomes continuous after this process.
4. Coordination by PCNA
Among the important factors that help in correct ligation is PCNA. It act as a sliding clamp. It keeps polymerase δ, FEN1 and ligase I in an ordered manner. When the synthesis and flap removal is finished PCNA allow the ligase to bind. This interaction also increases the activity of ligase. Without PCNA the joining of Okazaki fragments is very low.
In Prokaryotes
It is simpler in prokaryotic cells. DNA polymerase I remove the RNA primer and fill the gap. Only a single nick remain. This nick is sealed by DNA ligase. It is the same reaction where a phosphodiester bond is formed to give a continuous DNA strand.
Differences of prokaryotes and eukaryotes Okazaki fragments
Structural Differences
- The fragment length in prokaryotes is long, usually around 1000–2000 nucleotides.
- In eukaryotes the fragments are much shorter and are about 100–200 nucleotides.
- It is observed that prokaryotes do not have chromatin structure, so the fragment size is mainly controlled by replisome activity.
- In eukaryotes the small fragment length is related with the nucleosome core (about 150 bp) which act as a barrier during synthesis.
- The prokaryotic DNA is single and circular and replication occur in the cytoplasm.
- The eukaryotic DNA is linear and arranged in nucleus, and replication occur in the nucleus only.
- Only one origin of replication is present in prokaryotes.
- Many origins are present in eukaryotes and thousands of replicons can be formed.
- The rate of replication is high in prokaryotes and polymerase III can add nucleotides very fast.
- The eukaryotic synthesis is slow and only about 100 nucleotides per second is added.
Enzymatic and Maturation Differences
- For elongation–
- In prokaryotes DNA polymerase III is the main enzyme for extending Okazaki fragments.
- In eukaryotes polymerase δ is used for lagging strand synthesis and polymerase α/primase initiate the fragment formation.
- For primer removal and filling–
- The prokaryotic system is simple. DNA polymerase I remove the RNA primer by its 5′→3′ exonuclease activity and then fill the gap.
- After this the remaining nick is sealed by DNA ligase.
- The eukaryotic process is more complex. It is coordinated by PCNA sliding clamp.
- Polymerase δ displace the primer forming a flap and this flap is cut by FEN1 enzyme.
- Sometimes a long flap can be formed where DNA2 helicase/nuclease can be involved mainly in yeast.
- The major pathway in higher eukaryotes is the short flap pathway which depend on FEN1.
- Finally DNA ligase I seal the nick.
- Genome maintenance–
- The eukaryotic maturation need many enzymes and steps, so it is more error-sensitive and require strict regulation.
- The prokaryotic system is simple, as only polymerase I and ligase perform the full reaction.
Differences Between Prokaryotic and Eukaryotic Okazaki Fragments
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Fragment length | The fragments are long and usually about 1000–2000 nucleotides. | The fragments are short and mostly around 100–200 nucleotides. |
| Reason for fragment size | It is not restricted by chromatin because the DNA is free and simple. Replisome activity mainly decide the size. | The fragment length match with the nucleosome core and linker region, so the chromatin act as a barrier. |
| DNA organization | It has a single circular chromosome and replication take place in cytoplasm. | It has many linear chromosomes arranged in nucleus and replication occur inside the nucleus. |
| Replication origins | Only one origin of replication is present. | Many origins are present and thousands of replicons can be formed. |
| Rate of synthesis | It is very fast. Polymerase III add nucleotides very quickly. | It is slower. About 100 nucleotides is added per second. |
| Main enzyme for synthesis | DNA polymerase III extend the Okazaki fragments. | Polymerase δ synthesize the lagging strand and polymerase α/primase form the initial segment. |
| Primer removal | DNA polymerase I remove the RNA primer by its 5′→3′ exonuclease activity and fill the gap. | Polymerase δ displace the primer forming flap and FEN1 cut the flap. |
| Maturation pathway | It is simple and called nick translation. Only polymerase I and DNA ligase complete the process. | It is complex. PCNA coordinate the reaction. FEN1 is the major nuclease. In some cases DNA2 can act in long flap formation (mainly in yeast). |
| Ligation step | DNA ligase seal the final nick. | DNA ligase I is used to seal the nick. |
| Genome maintenance | It require less regulation because the process is simple. | It need strict control since many steps and enzymes are involved. |
MCQ Quiz
What are Okazaki fragments primarily associated with?
a) Transcription of RNA
b) Replication of the leading strand of DNA
c) Replication of the lagging strand of DNA
d) Translation of proteins
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: c) Replication of the lagging strand of DNA
[/expand]
Which enzyme is responsible for synthesizing the RNA primers that initiate Okazaki fragments?
a) DNA polymerase
b) DNA ligase
c) Primase
d) Helicase
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: c) Primase
[/expand]
Which enzyme is responsible for joining Okazaki fragments together?
a) DNA polymerase
b) DNA ligase
c) Primase
d) Helicase
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: b) DNA ligase
[/expand]
On which strand of the DNA do Okazaki fragments form?
a) Leading strand
b) Lagging strand
c) Both strands
d) Neither strand
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: b) Lagging strand
[/expand]
Why do Okazaki fragments form during DNA replication?
a) Because DNA is single-stranded
b) Due to the antiparallel nature of DNA and the directionality of DNA polymerase
c) To repair damaged DNA
d) To assist in DNA transcription
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: b) Due to the antiparallel nature of DNA and the directionality of DNA polymerase
[/expand]
Which of the following enzymes is NOT directly involved in the processing of Okazaki fragments?
a) DNA ligase
b) Primase
c) RNA polymerase
d) DNA polymerase
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: c) RNA polymerase
[/expand]
Approximately how long are Okazaki fragments in eukaryotes?
a) 10-50 nucleotides
b) 100-200 nucleotides
c) 500-1000 nucleotides
d) 2000-5000 nucleotides
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: b) 100-200 nucleotides
[/expand]
Which enzyme unwinds the DNA double helix, creating a need for Okazaki fragments on the lagging strand?
a) DNA ligase
b) DNA gyrase
c) DNA helicase
d) DNA topoisomerase
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: c) DNA helicase
[/expand]
After the RNA primers of Okazaki fragments are removed, they are replaced by:
a) Proteins
b) Lipids
c) DNA
d) RNA
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: c) DNA
[/expand]
Which of the following best describes the synthesis of the lagging strand during DNA replication?
a) Continuous and smooth
b) Discontinuous and in fragments
c) In a 3′ to 5′ direction
d) Without the need for primers
[expand title=”Show answer” swaptitle=”Hide answer”]
Answer: b) Discontinuous and in fragments
[/expand]
FAQ
What are the Okazaki fragments?
Okazaki fragments are short sequences of DNA nucleotides synthesized discontinuously on the lagging strand during DNA replication.
What is the function of the Okazaki fragments?
The function of Okazaki fragments is to enable the synthesis of the lagging strand of DNA in the 5′ to 3′ direction, consistent with the directionality of DNA polymerase.
What are Okazaki fragments and why are they formed?
Okazaki fragments are short DNA sequences formed on the lagging strand during DNA replication. They are formed due to the antiparallel nature of DNA and the directionality of DNA polymerase, which can only synthesize DNA in the 5′ to 3′ direction.
Why Okazaki fragments are formed?
They are formed to facilitate the synthesis of the lagging strand in the 5′ to 3′ direction during DNA replication.
Where are Okazaki fragments found?
Okazaki fragments are found on the lagging strand of the DNA during replication.
Why Okazaki fragments are discontinuous?
Okazaki fragments are discontinuous because the lagging strand is synthesized in short segments, opposite to the direction of the replication fork.
Which Okazaki fragment was made first?
The Okazaki fragment closest to the replication origin was made first.
What are the Okazaki fragments in DNA chain growth?
In DNA chain growth, Okazaki fragments represent the short, discontinuous segments of DNA synthesized on the lagging strand.
Are Okazaki fragments important?
Yes, Okazaki fragments are crucial for the accurate replication of the lagging strand of DNA.
How many Okazaki fragments are there?
The number of Okazaki fragments varies depending on the length of the lagging strand being replicated. There can be hundreds to thousands of Okazaki fragments formed during the replication of a single DNA molecule.
Is an Okazaki fragment DNA or RNA?
An Okazaki fragment is DNA, but it initially starts with an RNA primer which is later replaced by DNA.
- Alberts, B. M., Barry, K., Bedinger, P., Formosa, T., Jongeneel, C. V., & Kreuzer, K. N. (1982). Studies on DNA replication in the T4 bacteriophage in vitro system. Cold Spring Harbor Symposia on Quantitative Biology, 47, 655–668. https://doi.org/10.1101/sqb.1982.047.01.077
- Ahn, B., Harrigan, J. A., Indig, F. E., Wilson, D. M., III, & Bohr, V. A. (2004). Regulation of WRN helicase activity in human base excision repair. Journal of Biological Chemistry, 279(51), 53465–53474. https://doi.org/10.1074/jbc.M406206200
- Araujo, S. J., Tirode, F., Coin, F., Pospiech, H., Syvaoja, J. E., Stucki, M., Hubscher, U., Egly, J. M., & Wood, R. D. (2000). Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH, and modulation by CAK. Genes & Development, 14(3), 349–359.
- Bae, S. H., Bae, K. H., Kim, J. A., & Seo, Y. S. (2001). RPA governs endonuclease switching during processing of Okazaki fragments in eukaryotes. Nature, 412(6845), 456–461. https://doi.org/10.1038/35086586
- Balakrishnan, L., & Bambara, R. A. (2011). Eukaryotic lagging strand DNA replication employs a multi-pathway mechanism that protects genome integrity. Journal of Biological Chemistry, 286(8), 6865–6870. https://doi.org/10.1074/jbc.R110.209502
- Balakrishnan, L., & Bambara, R. A. (2013). Okazaki fragment metabolism. Cold Spring Harbor Perspectives in Biology, 5(2), a010173. https://doi.org/10.1101/cshperspect.a010173
- Balakrishnan, L., Brandt, P. D., Lindsey-Boltz, L. A., Sancar, A., & Bambara, R. A. (2009). Long patch base excision repair proceeds via coordinated stimulation of the multienzyme DNA repair complex. Journal of Biological Chemistry, 284(23), 15158–15172. https://doi.org/10.1074/jbc.M109.029413
- Balakrishnan, L., Stewart, J., Polaczek, P., Campbell, J. L., & Bambara, R. A. (2010). Acetylation of Dna2 endonuclease/helicase and flap endonuclease 1 by p300 promotes DNA stability by creating long flap intermediates. Journal of Biological Chemistry, 285(6), 4398–4404. https://doi.org/10.1074/jbc.M109.024828
- Bambara, R. A., Murante, R. S., & Henricksen, L. A. (1997). Enzymes and reactions at the eukaryotic DNA replication fork. Journal of Biological Chemistry, 272(8), 4647–4650.
- Bhattacharyya, A., Ear, U. S., Koller, B. H., Weichselbaum, R. R., & Bishop, D. K. (2000). The breast cancer susceptibility gene BRCA1 is required for subnuclear assembly of Rad51 and survival following treatment with the DNA cross-linking agent cisplatin. Journal of Biological Chemistry, 275(31), 23899–23903. https://doi.org/10.1074/jbc.M002773200
- Biswas, E. E., Zhu, F. X., & Biswas, S. B. (1997). Stimulation of RTH1 nuclease of the yeast Saccharomyces cerevisiae by replication protein A. Biochemistry, 36(20), 5955–5962. https://doi.org/10.1021/bi962916u
- Brosh, R. M., Jr., von Kobbe, C., Sommers, J. A., Karmakar, P., Opresko, P. L., Piotrowski, J., Dianova, I., Dianov, G. L., & Bohr, V. A. (2001). Werner syndrome protein interacts with human flap endonuclease 1 and stimulates its cleavage activity. EMBO Journal, 20(20), 5791–5801. https://doi.org/10.1093/emboj/20.20.5791
- Burgers, P. M. J. (2009). Polymerase dynamics at the eukaryotic DNA replication fork. Journal of Biological Chemistry, 284(7), 4041–4045. https://doi.org/10.1074/jbc.R800062200
- Caglayan, M., El-Khamisy, S. F., & Wilson, S. H. (2014). Role of polymerase beta in complementing aprataxin deficiency during abasic-site base excision repair. Nature Structural & Molecular Biology, 21(5), 497–499. https://doi.org/10.1038/nsmb.2818
- Caglayan, M., El-Khamisy, S. F., & Wilson, S. H. (2015). Complementation of aprataxin deficiency by base excision repair enzymes. Nucleic Acids Research, 43(4), 2271–2281. https://doi.org/10.1093/nar/gkv079
- Chai, Q., Zheng, L., Zhou, M., Turchi, J. J., & Shen, B. (2003). Interaction and stimulation of human FEN-1 nuclease activities by heterogeneous nuclear ribonucleoprotein A1 in alpha-segment processing during Okazaki fragment maturation. Biochemistry, 42(49), 15045–15052. https://doi.org/10.1021/bi035249p
- Chapados, B. R., Hosfield, D. J., Han, S., Qiu, J., Yelent, B., Shen, B., & Tainer, J. A. (2004). Structural basis for FEN-1 substrate specificity and PCNA-mediated activation in DNA replication and repair. Cell, 116(1), 39–50. https://doi.org/10.1016/s0092-8674(03)01036-5
- Clausen, A. R., Lujan, S. A., Burkholder, A. B., Orebaugh, C. D., Williams, J. S., Clausen, M. F., Kunkel, T. A. (2015). Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nature Structural & Molecular Biology, 22(3), 185–191.
- Dianova, I. I., Bohr, V. A., & Dianov, G. L. (2001). Interaction of human AP endonuclease 1 with flap endonuclease 1 and proliferating cell nuclear antigen involved in long-patch base excision. Biochemistry, 40(42), 12639–12644. https://doi.org/10.1021/bi011319n
- Duxin, J. P., Dao, B., Martinsson, P., Rajala, N., Guittat, L., Campbell, J. L., Spelbrink, J. N., & Stewart, S. A. (2009). Human Dna2 is a nuclear and mitochondrial DNA maintenance protein. Molecular and Cellular Biology, 29(15), 4274–4282. https://doi.org/10.1128/MCB.00512-09
- Frank, G., Qiu, J., Zheng, L., & Shen, B. (2001). Stimulation of eukaryotic flap endonuclease-1 activities by proliferating cell nuclear antigen (PCNA) is independent of its in vitro interaction via a consensus PCNA binding region. Journal of Biological Chemistry, 276(39), 36295–36302. https://doi.org/10.1074/jbc.M104712200
- Gary, R., Park, M. S., Nolan, J. P., Cornelius, H. L., Kozyreva, O. G., Tran, H. T., Lobachev, K. S., Resnick, M. A., & Gordenin, D. A. (1999). A novel role in DNA metabolism for the binding of Fen1/Rad27 to PCNA and implications for genetic risk. Molecular and Cellular Biology, 19(8), 5373–5382. https://doi.org/10.1128/MCB.19.8.5373
- Harrington, J. J., & Lieber, M. R. (1994). The characterization of a mammalian structure-specific DNA endonuclease. EMBO Journal, 13(5), 1235–1246.
- Hasan, S., Stucki, M., Hassa, P. O., Imhof, R., Gehrig, P., Hunziker, P., Hubscher, U., & Hottiger, M. O. (2001). Regulation of human flap endonuclease-1 activity by acetylation through the transcriptional coactivator p300. Molecular Cell, 7(6), 1221–1231. https://doi.org/10.1016/s1097-2765(01)00262-1
- Kao, H. I., & Bambara, R. A. (2003). The protein components and mechanism of eukaryotic Okazaki fragment maturation. Critical Reviews in Biochemistry and Molecular Biology, 38(5), 433–452. https://doi.org/10.1080/713608753
- Klungland, A., & Lindahl, T. (1997). Second pathway for completion of human DNA base excision-repair: reconstitution with purified proteins and requirement for DNase IV (FEN1). EMBO Journal, 16(11), 3341–3348. https://doi.org/10.1093/emboj/16.11.3341
- Kornberg, A., & Baker, T. A. (1992). DNA replication. W.H. Freeman.
Kucherlapati, M., Yang, K., Kuraguchi, M., Zhao, J., Lia, M., Heyer, J., Kane, M. F., Fan, K., Russell, R., Brown, A. M. C., Kneitz, W., Edelmann, R., Kolodner, M. R., Lipkin, R., & Kucherlapati, R. (2002). Haploinsufficiency of Flap endonuclease (Fen1) leads to rapid tumor progression. Proceedings of the National Academy of Sciences of the United States of America, 99(15), 9924–9929. https://doi.org/10.1073/pnas.152342599 - Li, X., Li, J., Harrington, J., Lieber, M. R., & Burgers, P. M. J. (1995). Lagging strand DNA synthesis at the eukaryotic replication fork involves binding and stimulation of FEN-1 by proliferating cell nuclear antigen. Journal of Biological Chemistry, 270(37), 22109–22112. https://doi.org/10.1074/jbc.270.37.22109
- Lieber, M. R. (1997). The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays, 19(3), 233–240. https://doi.org/10.1002/bies.970190308
- Lindahl, T. (1993). Instability and decay of the primary structure of DNA. Nature, 362(6422), 709–715. https://doi.org/10.1038/362709a0
- Liu, Y., Kao, H. I., & Bambara, R. A. (2004). Flap endonuclease 1: a central component of DNA metabolism. Annual Review of Biochemistry, 73, 589–615. https://doi.org/10.1146/annurev.biochem.73.012803.092453
- Liu, Y., Prasad, R., Beard, W. A., Hou, E. W., Horton, J. K., McMurray, C. T., & Wilson, S. H. (2009). Coordination between Pol {beta} and FEN1 can modulate CAG repeat expansion. Journal of Biological Chemistry, 284(41), 28352–28366.
- Mangiarini, L., Sathasivam, K., Seller, M., Cozens, B., Harper, A., Hetherington, C., Lawton, M., Trottier, Y., Lehrach, H., Davies, S. W., & Bates, G. P. (1996). Exon 1 of the HD gene with an expanded CAG repeat is sufficient to cause progressive neurological phenotype in transgenic mice. Cell, 87(3), 493–506. https://doi.org/10.1016/s0092-8674(00)81369-0
- Okazaki, R., Okazaki, T., Sakabe, K., Sugimoto, K., & Sugino, A. (1968). Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proceedings of the National Academy of Sciences of the United States of America, 59(2), 598–605. https://doi.org/10.1073/pnas.59.2.598
- Okazaki, T. (2017). Days weaving the lagging strand synthesis of DNA – A personal recollection of the discovery of Okazaki fragments and studies on discontinuous replication mechanism. Proceedings of the Japan Academy. Series B, Physical and Biological Sciences, 93(5), 322–338. https://doi.org/10.2183/pjab.93.020
- Parrish, J. Z., Yang, C., Shen, B., & Xue, D. (2003). CRN-1, a Caenorhabditis elegans FEN-1 homologue, cooperates with CPS-6/EndoG to promote apoptotic DNA degradation. EMBO Journal, 22(13), 3451–3460. https://doi.org/10.1093/emboj/cdg319