What is a Nucleotide?
A nucleotide is a fundamental component of nucleic acids, such as DNA and RNA. It is composed of three main parts: a pentose sugar, a nitrogenous base, and a phosphate group. Nucleotides are often referred to as the building blocks of DNA due to their crucial role in forming the DNA molecule.
The nitrogenous bases found in nucleotides can be categorized into two types: purines and pyrimidines. Purines include adenine (A) and guanine (G), while pyrimidines consist of cytosine (C), thymine (T) in DNA (or uracil (U) in RNA). These nitrogenous bases are derived from specific parent compounds.
When examining the nucleotides present in DNA, they contain a 2′-deoxy-D-ribose sugar. The 2′-deoxyribose differs from D-ribose (found in RNA) in that it lacks an alcohol group (-OH) at the second position of the pentose structure. This absence of an alcohol group distinguishes 2′-deoxyribose from D-ribose, leading to its name.
On the other hand, D-ribose pentose, which is found in RNA nucleotides, possesses an -OH group at the second position of the pentose structure. This presence of the -OH group is a key distinction between the sugar component of DNA and RNA nucleotides.
In both DNA and RNA, the pentose sugars exist in their β-furanose form, which refers to a close five-membered ring structure. This β-furanose conformation plays a significant role in the stability and structure of nucleic acids.
Overall, a nucleotide is a vital molecular unit that combines a pentose sugar, a nitrogenous base, and a phosphate group. These nucleotides link together to form the DNA double helix or RNA single strands, carrying the genetic information necessary for cellular processes and heredity.
Characteristics of Nucleotide
Nucleotides possess several characteristic features that contribute to their structural and functional roles:
- Covalent Bonding: The nitrogenous base of a nucleotide is covalently linked to the pentose sugar via an N-glycosidic bond. In pyrimidines, this bond occurs at the N-1 position, while in purines, it forms at the N-9 position. This covalent bond ensures a stable connection between the base and sugar components.
- Purine and Pyrimidine Bases: The major purine bases found in DNA and RNA are Adenosine (A) and Guanine (G). These bases contain a double-ring structure. In DNA, the major pyrimidine bases are Thymine (T) and Cytosine (C), each with a single-ring structure. However, in RNA, Thymine is replaced by Uracil (U) as a major pyrimidine base.
- Phosphodiester Linkages: Consecutive nucleotides within a DNA or RNA strand are linked by phospho-diester bonds. Specifically, the 5′ phosphate group of one nucleotide forms a bond with the 3′ hydroxyl group of the adjacent nucleotide. This linkage creates a continuous chain of nucleotides along the DNA or RNA strand.
- Hydrogen Bonding: Complementary nucleotides in DNA are connected by hydrogen bonds. Adenosine (A) forms two hydrogen bonds with Thymine (T), while Guanine (G) forms three hydrogen bonds with Cytosine (C). These hydrogen bonds contribute to the stability and specificity of base pairing within the DNA double helix.
- Backbone Structure: The backbone of DNA is composed of a linear chain of nucleotides. The phosphate groups of successive nucleotides form a continuous backbone, with the sugar-phosphate backbone running in an alternating pattern. This backbone provides structural support and stability to the DNA molecule.
In summary, nucleotides exhibit characteristic features such as covalent bonding between the nitrogenous base and pentose sugar, specific purine and pyrimidine bases, phosphodiester linkages between consecutive nucleotides, hydrogen bonding between complementary bases, and the formation of a nucleotide chain that constitutes the backbone of DNA. These characteristics contribute to the unique structure and function of nucleic acids.
Biosynthesis of Nucleotides
The nucleotides are synthesized by two pathways:
- De novo synthesis
- Salvage pathway
A. De novo synthesis of nucleotides
De novo synthesis of nucleotides is the process by which cells generate new nucleotides from simple precursor molecules. Nucleotides are the building blocks of DNA and RNA, and they are also involved in numerous other cellular processes, such as energy metabolism, signaling, and enzyme regulation.
In de novo synthesis, nucleotides are synthesized from basic precursor molecules, including amino acids, sugars, and various other small molecules. The process involves several enzymatic reactions, each catalyzed by specific enzymes. These reactions are tightly regulated and occur in multiple steps.
The de novo synthesis pathway differs slightly for purine and pyrimidine nucleotides:
- De novo synthesis of purine nucleotides: Purine nucleotides, such as adenosine triphosphate (ATP) and guanosine triphosphate (GTP), are synthesized from precursor molecules like amino acids and ribose-5-phosphate. The pathway starts with the formation of inosine monophosphate (IMP), which serves as a precursor for AMP (adenosine monophosphate) and GMP (guanosine monophosphate). The synthesis of purine nucleotides involves several enzymatic reactions and requires the input of energy in the form of ATP and GTP.
- De novo synthesis of pyrimidine nucleotides: Pyrimidine nucleotides, including cytidine triphosphate (CTP) and uridine triphosphate (UTP), are synthesized from simpler molecules like aspartate, glutamine, and bicarbonate. The pathway starts with the formation of carbamoyl phosphate, which combines with aspartate to produce orotate. Orotate is then converted into the common precursor molecule, uridine monophosphate (UMP). UMP can be further modified to generate other pyrimidine nucleotides.
The de novo synthesis of nucleotides is crucial for cell growth, development, and maintenance. It ensures an adequate supply of nucleotides for DNA and RNA synthesis, as well as for various cellular processes requiring nucleotide cofactors or signaling molecules. The pathway is tightly regulated to balance the cellular demand for nucleotides with the available resources.
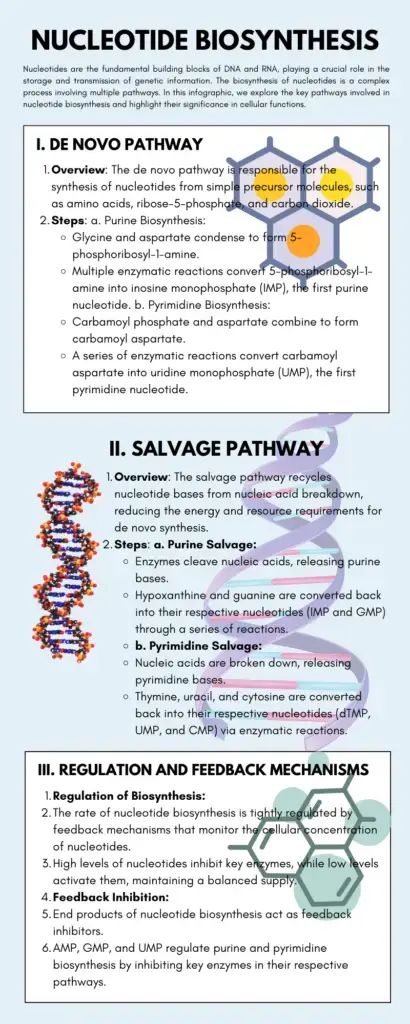
Synthesis of Pyrimidine nucleotides
The synthesis of pyrimidine nucleotides involves two major steps: the formation of a pyrimidine ring and the attachment of the pentose sugar (ribose-5-phosphate) to the ring structure. Here is a detailed explanation of the synthesis process:
- Formation of the pyrimidine ring: a. The synthesis begins with the conversion of aspartate and carbamoyl phosphate to N-carbamoyl aspartate. This reaction is catalyzed by the enzyme aspartate transcarbamoylase (ATCase). b. N-carbamoyl aspartate undergoes a reaction where one water molecule is removed, resulting in the formation of L-dihydroorotate. This step is facilitated by the enzyme dihydroorotase. c. L-dihydroorotate is then converted to orotate in the presence of dihydroorotate dehydrogenase. d. Orotate is the key compound to which the ribose-5-phosphate sugar is added in the subsequent step.
- Attachment of the pentose sugar: a. Orotate reacts with ribose-5-phosphate, which is provided by phosphoribosyl pyrophosphate (PRPP), in the presence of orotate phosphoribosyltransferase. This reaction forms orotidylate (orotidylic acid). b. Orotidylate undergoes decarboxylation, catalyzed by orotidylate decarboxylase, to form uridylate (UMP), also known as uridine 5′-monophosphate. c. Uridylate is further phosphorylated in subsequent steps to form uridine 5′-triphosphate (UTP). d. Cytidine 5′-triphosphate (CTP) is formed from UTP through the action of the enzyme cytidine synthetase, with glutamine serving as the nitrogen donor.
By following these steps, the cell synthesizes the pyrimidine nucleotides uridine 5′-triphosphate (UTP) and cytidine 5′-triphosphate (CTP). These nucleotides are essential for DNA and RNA synthesis, as well as other cellular processes. The synthesis of pyrimidines requires the involvement of various enzymes and the availability of substrates such as aspartate, PRPP, carbamoyl phosphate, glutamine, ATP, and CO2. Carbamoyl phosphate, a critical component in pyrimidine biosynthesis, is produced in the cytosol by the enzyme carbamoyl phosphate synthetase II.
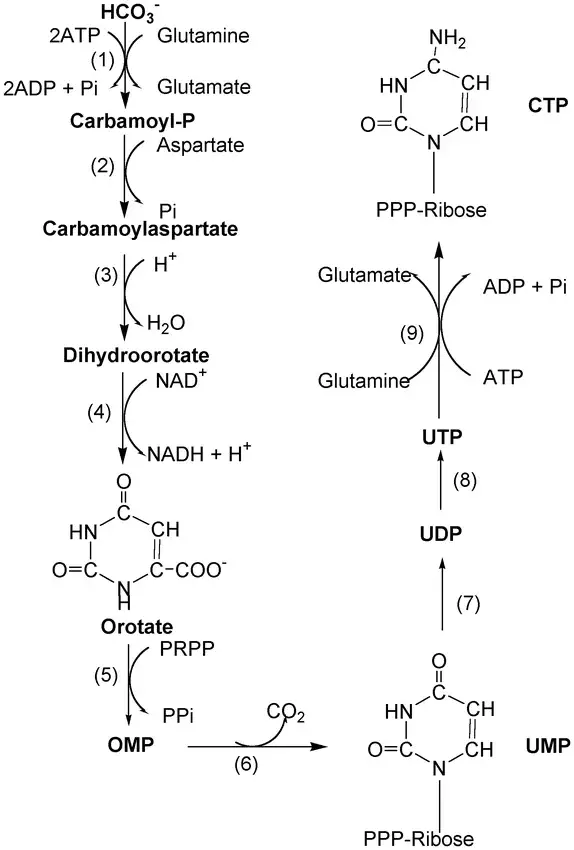
Synthesis of Purine nucleotides
The synthesis of purine nucleotides follows a different pathway compared to pyrimidine nucleotides. Here is a detailed explanation of the synthesis process:
- Initiation of purine synthesis:
- The synthesis of purine nucleotides begins with phosphoribosyl pyrophosphate (PRPP). Unlike pyrimidine synthesis, where the ring is formed first, in purine synthesis, the purine ring is built around the sugar (PRPP).
- Formation of Inosinate (IMP):
- a. PRPP reacts with glutamine in the presence of the enzyme glutamine phosphoribosylpyrophosphate amidotransferase (Glutamine-PRPP) to form phosphoribosyl amine (PRA). The amino group from glutamine is attached to the C1 position of PRPP.
- b. PRA is converted to glycinamide ribonucleotide (GAR) by the enzyme glycinamide ribonucleotide synthase (GAR synthase). Three atoms from a glycine molecule are added to the nitrogen atom of PRA.
- c. GAR is transformed into formylglycinamide ribonucleotide (FGAR) by the enzyme glycinamide ribotide transformylase (GAR transformylase). The added glycine molecule is formylated using N10-formyl tetrahydrofolate.
- d. FGAR is further converted to formylglycinamidine (FGAM) by the enzyme formylglycinamide synthase (FGAM synthase).
- e. FGAM is catalyzed by aminoimidazole ribotide synthase to form aminoimidazole ribonucleotide (AIR), which is then converted to carboxyaminoimidazole ribonucleotide (CAIR) by aminoimidazole ribotide carboxylase (AIR carboxylase).
- f. CAIR reacts with aspartate in the presence of succinylaminoimidazolecarboxamide ribotide synthase to form succinylaminoimidazolecarboxamide ribotide (SAICAR).
- g. SAICAR is converted to aminoimidazole carboxamide ribonucleotide (AICAR) by the enzyme adenylosuccinate lyase (ASL).
- h. AICAR is transformed into formylaminoimidazole carboxamide ribonucleotide (FAICAR) by aminoimidazole carboxamide ribotide transformylase.
- i. Finally, the second ring of the purine is closed by the enzyme IMP cyclohydrolase, resulting in the formation of inosinate (IMP).
- Synthesis of AMP and GMP:
- From IMP, further modifications occur to synthesize adenosine monophosphate (AMP) and guanosine monophosphate (GMP).
- Enzymes involved in this process include adenylosuccinate synthetase and adenylosuccinate lyase (for AMP synthesis), as well as IMP dehydrogenase and GMP synthetase (for GMP synthesis).
By following these steps, the cell synthesizes the purine nucleotide inosinate (IMP), which serves as the precursor for the synthesis of adenosine monophosphate (AMP) and guanosine monophosphate (GMP). The synthesis of purine nucleotides requires the involvement of various enzymes, as mentioned above, and the availability of substrates such as PRPP, glutamine, glycine, aspartate, and N10-formyl tetrahydrofolate.
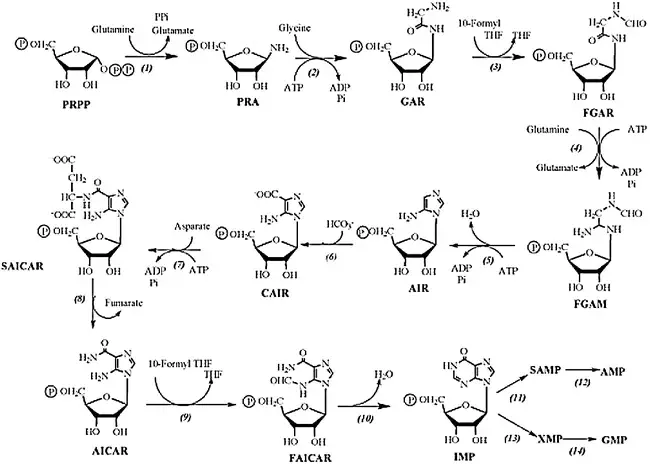
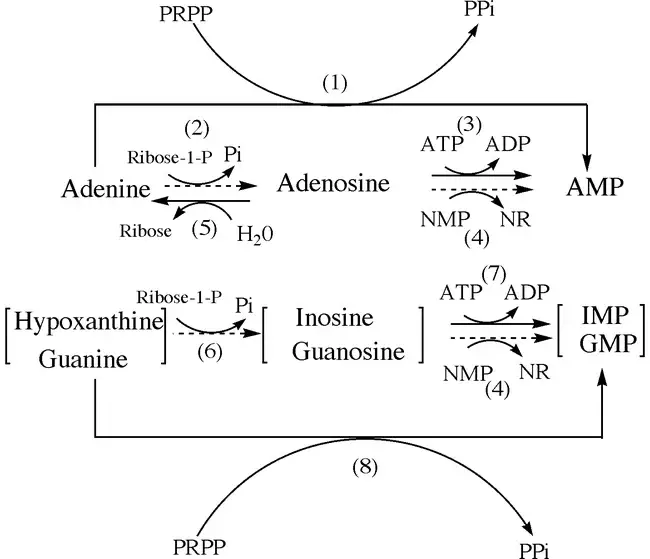
B. Salvage pathway of Purine nucleotides
The salvage pathway of purine nucleotides is an alternative pathway for the synthesis of purine nucleotides using pre-existing purine bases and nucleosides. Instead of de novo synthesis, the salvage pathway recycles and converts purine bases and nucleosides back into nucleotides. Here is an explanation of the salvage pathway for purine nucleotides:
- Adenine salvage:
- Adenine, a purine base, can be salvaged and converted into adenosine monophosphate (AMP) through the action of the enzyme adenosine phosphoribosyltransferase (APRT). APRT catalyzes the transfer of a phosphoribosyl group from phosphoribosyl pyrophosphate (PRPP) to adenine, forming adenosine monophosphate (AMP).
- Guanine and hypoxanthine salvage:
- Guanine and hypoxanthine, two other purine bases, are salvaged by the enzyme hypoxanthine-guanine phosphoribosyltransferase (HGPRT) through a process called phosphoribosylation. HGPRT catalyzes the transfer of a phosphoribosyl group from PRPP to guanine or hypoxanthine, forming guanosine monophosphate (GMP) and inosine monophosphate (IMP), respectively.
- Adenosine deamination:
- Adenosine, a purine nucleoside, can undergo deamination by the enzyme adenosine deaminase. This deamination reaction converts adenosine to inosine, which can then be salvaged through the hypoxanthine salvage pathway mentioned above. Therefore, adenosine can be converted to hypoxanthine and incorporated into the purine nucleotide pool via the salvage pathway.
The salvage pathway of purine nucleotides is an important mechanism for recycling and reutilizing purine bases and nucleosides, thereby conserving energy and resources. It allows the cell to maintain a balanced supply of purine nucleotides without solely relying on de novo synthesis. The enzymes APRT and HGPRT play key roles in salvaging and incorporating purine bases into the purine nucleotide pool, contributing to the overall cellular metabolism and nucleotide homeostasis.
Salvage pathway of Pyrimidine nucleotides (Thymine)
The salvage pathway of pyrimidine nucleotides, specifically for thymine, involves the enzyme thymidine kinase. Thymidine kinase is responsible for salvaging deoxythymidine triphosphate (dTTP) from thymidine or deoxyuridine. Here is an explanation of the salvage pathway for pyrimidine nucleotides, focusing on thymine:
- Thymidine salvage:
- Thymidine, a pyrimidine nucleoside, can be salvaged and converted into deoxythymidine triphosphate (dTTP) through the action of the enzyme thymidine kinase.
- Thymidine kinase catalyzes the transfer of a phosphate group from ATP to thymidine, resulting in the formation of thymidine monophosphate (dTMP).
- dTMP is further phosphorylated by additional kinases to form dTDP (deoxythymidine diphosphate) and finally dTTP (deoxythymidine triphosphate). dTTP is an essential building block for DNA synthesis and serves as a precursor for the incorporation of thymine into DNA strands.
- Deoxyuridine salvage:
- In addition to thymidine, thymidine kinase can also salvage deoxyuridine, another pyrimidine nucleoside.
- Deoxyuridine is phosphorylated by thymidine kinase, resulting in the formation of deoxyuridine monophosphate (dUMP).
- dUMP can then undergo further phosphorylation by other kinases to form dTDP and dTTP, similar to the thymidine salvage pathway described above.
By salvaging thymidine and deoxyuridine, thymidine kinase plays a crucial role in recycling and reutilizing these pyrimidine nucleosides, allowing the cell to maintain a balanced supply of pyrimidine nucleotides for DNA synthesis. The salvage pathway conserves energy and resources by reusing existing nucleosides instead of relying solely on de novo synthesis. The enzyme thymidine kinase and its activity in the salvage pathway contribute to the overall metabolism and homeostasis of pyrimidine nucleotides in the cell.
Pyrimidine biosynthesis regulation
The biosynthesis of pyrimidines is tightly regulated to ensure the proper balance of nucleotide production within the cell. The regulation occurs at various levels, primarily involving the enzyme Aspartate transcarbamoylase (ATCase) and the availability of key metabolic intermediates. Here is an explanation of the regulation of pyrimidine biosynthesis:
- Regulation by ATCase:
- ATCase is a multi-subunit allosteric enzyme that catalyzes the conversion of aspartate to carbamoyl aspartate, a key step in pyrimidine biosynthesis.
- ATP acts as an activator of ATCase, promoting the synthesis of pyrimidines.
- On the other hand, CTP acts as an inhibitor of ATCase, exerting feedback inhibition. When CTP levels are high, CTP binds to ATCase, inhibiting its activity and thus slowing down the synthesis of pyrimidines.
- This feedback inhibition by CTP is an important mechanism to regulate pyrimidine biosynthesis, as it ensures that the production of pyrimidine nucleotides is controlled when sufficient levels of CTP are present in the cell.
- Regulation at PRPP Formation:
- The availability of phosphoribosyl pyrophosphate (PRPP), a precursor molecule for nucleotide synthesis, also plays a role in regulating pyrimidine biosynthesis.
- High levels of PRPP indicate a high demand for nucleotide synthesis, including pyrimidines. Therefore, high levels of PRPP can serve as an indicator for increased pyrimidine synthesis.
- CTP synthetase, an enzyme involved in the conversion of uridine triphosphate (UTP) to CTP, is feedback inhibited by CTP. When CTP levels are high, it binds to CTP synthetase, reducing its activity and thus modulating the production of CTP.
- Additionally, the activity of CTP synthetase can be activated by another nucleotide, guanosine triphosphate (GTP), providing a mechanism for adjusting the production of CTP based on the availability of GTP.
By regulating the activity of ATCase and the availability of PRPP, the biosynthesis of pyrimidines can be finely tuned to meet the cellular requirements for nucleotide synthesis. The feedback inhibition by CTP ensures that the synthesis of pyrimidines is controlled when sufficient levels of end products are present, preventing overproduction. These regulatory mechanisms contribute to maintaining the balance of nucleotide metabolism and overall cellular homeostasis.
Purine biosynthesis regulation
The biosynthesis of purines is tightly regulated at multiple levels to maintain the balance of nucleotide production within the cell. Regulation occurs at the enzymes involved in the pathway, particularly PRPP synthetase, PRPP amidotransferase, and the branching point of inosine monophosphate (IMP). Here is an explanation of the regulation of purine biosynthesis:
- Regulation by PRPP Synthetase:
- PRPP synthetase is the first regulatory point in the purine biosynthesis pathway. It catalyzes the formation of phosphoribosyl pyrophosphate (PRPP), a key precursor in nucleotide synthesis.
- Feedback inhibition is exerted by the end products of purine metabolism, adenosine monophosphate (AMP) and guanosine monophosphate (GMP), on PRPP synthetase.
- When AMP and GMP levels are high, they bind to PRPP synthetase, inhibiting its activity and reducing the production of PRPP. This feedback inhibition ensures that the synthesis of purine nucleotides is controlled when sufficient levels of AMP and GMP are present in the cell.
- Regulation by PRPP Amidotransferase:
- PRPP amidotransferase is an allosteric enzyme that catalyzes the conversion of PRPP to 5-phosphoribosylamine, a key step in purine biosynthesis.
- Various nucleotides, including GTP, ATP, GDP, ADP, GMP, and AMP, bind to PRPP amidotransferase allosterically and regulate its activity.
- The binding of these nucleotides can either activate or inhibit the enzyme, depending on their concentrations. This allosteric regulation helps to fine-tune the biosynthesis of purines based on the cellular requirements.
- Regulation at the Branching Point of IMP:
- The last regulatory point in purine biosynthesis is the branching point of IMP. IMP serves as a precursor for the synthesis of both AMP and GMP.
- The relative amounts of ATP and GTP can influence the synthesis of AMP and GMP. Excess ATP accelerates the synthesis of GMP, while excess GTP accelerates the synthesis of AMP.
- This regulation ensures that the production of AMP and GMP is balanced according to the availability of their respective precursors, ATP and GTP.
By regulating the activity of PRPP synthetase, PRPP amidotransferase, and the branching point of IMP, the biosynthesis of purines can be controlled and coordinated with the cellular requirements for nucleotide synthesis. The feedback inhibition and allosteric regulation allow for precise control of purine metabolism, preventing excessive production and maintaining the balance of purine nucleotides in the cell.
Functions of Nucleotide
Nucleotides serve various important functions in the body and participate in numerous cellular processes. Here are some key functions of nucleotides:
- Energy Reserves: Nucleotides, particularly ATP (adenosine triphosphate), function as energy reserves in the body. ATP is often referred to as the “energy currency” of the cell. The hydrolysis of ATP releases a significant amount of energy that can be utilized by cells to perform various functions and drive metabolic processes.
- Phosphoryl Group Donors: ATP acts as a phosphoryl group donor in many cellular pathways. By transferring its phosphate groups to specific target molecules, ATP plays a crucial role in cellular processes such as signal transduction, enzyme activation, and biosynthesis.
- Co-factors for Enzymes: Nucleotides are essential components of co-factors for various enzymes. For example, Acetyl CoA, NAD+ (Nicotinamide adenine dinucleotide), and FAD (Flavin adenine dinucleotide) all contain nucleotide moieties. These co-factors are involved in key metabolic reactions and facilitate enzymatic activities, allowing for efficient biochemical transformations.
- Second Messengers: Some nucleotides function as second messengers, transmitting signals within cells. cAMP (adenosine cyclic monophosphate) is a common second messenger that is produced in response to hormones and various chemical signals. It plays a crucial role in signal transduction pathways, regulating cellular processes such as gene expression, metabolism, and ion channel activity. Similarly, cGMP (guanosine cyclic monophosphate) also serves as a second messenger, mediating various signaling pathways.
- Regulatory Functions: Nucleotides, such as ppGpp (guanosine tetraphosphate), have regulatory functions in specific cellular contexts. For example, in bacterial cells under amino acid starvation conditions, ppGpp is produced in response to the slowdown of protein production. It acts as a signaling molecule, controlling the expression of genes involved in adaptation and survival during periods of nutrient limitation.
In summary, nucleotides are essential molecules that play diverse roles in cellular processes. They provide energy, serve as co-factors for enzymes, act as second messengers, and participate in regulatory mechanisms. Their functions are crucial for maintaining cellular homeostasis and supporting various physiological processes in the body.
FAQ
What are nucleotide biosynthesis pathways?
Nucleotide biosynthesis pathways refer to the series of biochemical reactions in cells that synthesize nucleotides, which are the building blocks of DNA and RNA.
Why are nucleotide biosynthesis pathways important in the body?
Nucleotide biosynthesis pathways are crucial for the maintenance and replication of genetic material, as well as for various cellular processes such as energy metabolism, signal transduction, and enzyme co-factor production.
What are the major differences between the de novo synthesis and salvage pathways of nucleotide biosynthesis?
De novo synthesis pathways involve the de novo (from scratch) production of nucleotides using simple precursors, while salvage pathways involve the recycling of pre-existing nucleotides from nucleic acid breakdown or dietary sources.
How are pyrimidine nucleotides synthesized in cells?
Pyrimidine nucleotides (cytosine, thymine, and uracil) are synthesized through a multistep process involving the formation of a pyrimidine ring and the attachment of a sugar molecule. Aspartate, carbamoyl phosphate, and PRPP (phosphoribosyl pyrophosphate) are key intermediates in pyrimidine biosynthesis.
How are purine nucleotides synthesized in cells?
Purine nucleotides (adenine and guanine) are synthesized through a complex series of reactions involving the formation of a purine ring. PRPP acts as the starting material, and various enzymes and co-factors are involved in the stepwise addition of atoms to form the complete purine ring structure.
What are the key regulatory steps in pyrimidine biosynthesis?
The key regulatory steps in pyrimidine biosynthesis include the enzyme Aspartate transcarbamoylase (ATCase), which is inhibited by CTP and activated by ATP. This feedback inhibition helps regulate the overall rate of pyrimidine synthesis.
How is the biosynthesis of purine nucleotides regulated?
The biosynthesis of purine nucleotides is regulated by feedback inhibition. PRPP synthetase, the first regulatory point in the pathway, is inhibited by AMP and GMP. Additionally, the branching point of IMP (inosine monophosphate) is regulated by ATP and GTP, which accelerate the synthesis of GMP and AMP, respectively.
What are the feedback inhibitors involved in the regulation of nucleotide biosynthesis?
Feedback inhibitors involved in the regulation of nucleotide biosynthesis include CTP, which inhibits Aspartate transcarbamoylase (ATCase) in pyrimidine biosynthesis, and AMP/GMP, which inhibit PRPP synthetase in purine biosynthesis.
How do nucleotide biosynthesis pathways contribute to cellular energy metabolism?
Nucleotide biosynthesis pathways contribute to cellular energy metabolism through the production of ATP, which serves as a universal energy currency in cells. ATP is synthesized during the biosynthesis of nucleotides and can be used to fuel various cellular processes.
Are there any diseases or disorders associated with abnormalities in nucleotide biosynthesis pathways?
Abnormalities in nucleotide biosynthesis pathways can lead to various diseases or disorders. For example, deficiencies in enzymes involved in purine or pyrimidine biosynthesis can result in metabolic disorders such as Lesch-Nyhan syndrome or Orotic aciduria, respectively. Understanding these pathways is important for diagnosing and treating such conditions.
References
- Moffatt BA, Ashihara H. Purine and pyrimidine nucleotide synthesis and metabolism. Arabidopsis Book. 2002;1:e0018. doi:10.1199/tab.0018.
- Zimmermann H. (2016). Extracellular ATP and other nucleotides-ubiquitous triggers of intercellular messenger release. Purinergic signalling, 12(1), 25–57. https://doi.org/10.1007/s11302-015-9483-2
- Lehninger Principles of Biochemistry – 6th ed- c2013
- Cell Biology, T Devasena, Oxford University Press.