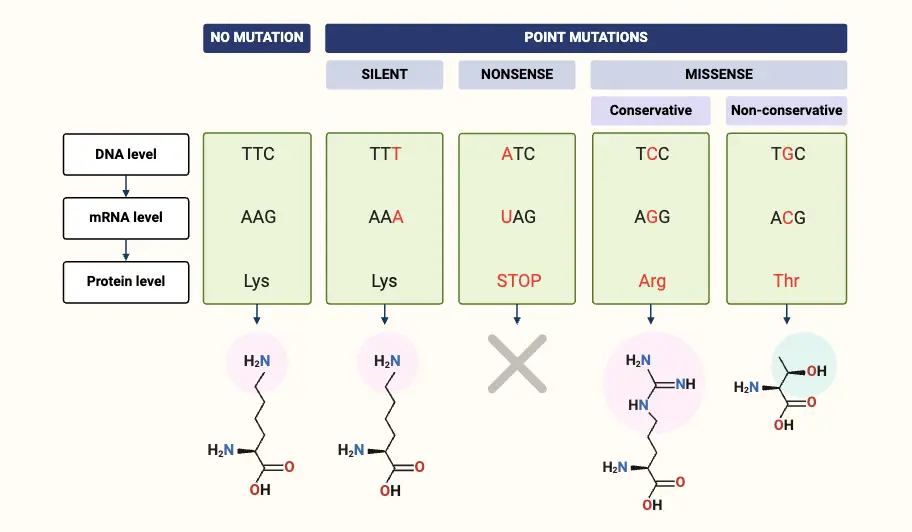
What is Nonsense Mutation?
- In genetics, a nonsense mutation is a specific type of point mutation that introduces a premature stop codon into the sequence of a gene. This mutation results in the translation of a truncated, often nonfunctional protein product because the translation machinery halts before completing the full-length protein.
- Nonsense mutations occur due to errors in DNA replication or repair processes, leading to the generation of a nonsense codon. These codons, which include UAA, UAG, and UGA in mRNA, do not specify an amino acid but instead signal the termination of protein synthesis. As a result, the polypeptide chain is cut short, leading to a protein that may be incomplete and dysfunctional.
- The functional impact of a nonsense mutation depends significantly on the position of the premature stop codon within the gene. If the stop codon appears early in the coding sequence, it typically results in a severely truncated protein that is often degraded by cellular proteasomes. This effect can be as severe as the complete deletion of the gene’s function.
- Conversely, if the nonsense mutation occurs closer to the end of the coding sequence, the protein may retain some of its original function. In rare instances, nonsense mutations can be beneficial if they produce a protein with enhanced functionality or altered properties that confer a selective advantage to the organism.
- Nonsense mutations are a type of loss-of-function mutation, which impairs the production of a specific protein. They are also referred to as chain termination mutations due to their role in halting protein synthesis prematurely. Diseases associated with nonsense mutations include Duchenne muscular dystrophy, cystic fibrosis, and various genetic disorders, illustrating their potential to disrupt critical biological functions.
- Overall, nonsense mutations represent a significant mechanism of genetic alteration with diverse impacts on protein function and organismal health.
Definition of Nonsense Mutation
A nonsense mutation is a genetic alteration that introduces a premature stop codon into the coding sequence of a gene, leading to the production of a truncated and often nonfunctional protein.
Causes of nonsense mutation
Nonsense mutations are a specific type of genetic mutation where a change in the DNA sequence introduces a premature stop codon into the coding sequence of a gene. This results in the early termination of protein synthesis and often leads to a truncated, nonfunctional protein. The causes of nonsense mutations can be classified into several key mechanisms:
- Nucleotide Substitution: One of the primary causes of nonsense mutations is the substitution of a nucleotide in the DNA sequence, converting a sense codon (which codes for an amino acid) into a stop codon. For instance, the replacement of a CGA codon with TGA or CAG with TAG can introduce a premature stop codon.
- Methylation-Mediated Deamination: Methylation-mediated deamination of cytosine residues is a common biochemical process that can result in nucleotide substitutions leading to nonsense mutations. This process involves the conversion of cytosine to uracil, which then pairs with adenine instead of guanine, causing a change in the codon sequence.
- Nonsense-Mediated mRNA Decay (NMD): Nonsense mutations often trigger the NMD pathway, which degrades the mRNA transcript containing the premature stop codon. This cellular mechanism prevents the translation of truncated proteins by eliminating faulty mRNA, thereby reducing potential harmful effects.
- Exon Skipping: In some cases, nonsense mutations can lead to exon skipping during the splicing of pre-mRNA. This occurs when the splicing machinery bypasses the exon containing the mutation, resulting in an altered mRNA transcript that may lack essential coding information.
- Impact on Splicing: Nonsense mutations can disrupt exonic splicing enhancers, which are sequences necessary for proper splicing of pre-mRNA. This disruption can lead to improper splicing events, such as exon skipping, which further impacts the mRNA’s stability and function.
- Gene-Specific Effects: The effects of nonsense mutations can vary depending on the specific gene involved. For example, tumor suppressor genes often exhibit nonsense mutations, which can contribute to cancer development, while oncogenes are more commonly affected by missense mutations.
- Position within Gene: The location of a nonsense mutation within a gene can influence the severity of the resulting phenotype. For example, in the dystrophin gene (DMD), the position of the nonsense mutation can determine the severity of muscular dystrophy.
- De Novo Mutations: Nonsense mutations can arise de novo, meaning they occur spontaneously in the germ cells and are not inherited from either parent. These de novo mutations can lead to genetic syndromes with distinct clinical features, such as developmental delays or congenital anomalies.
- Genetic Variation and Phenotype: The impact of a nonsense mutation on the phenotype often depends on the extent of the truncation and the functional domains of the protein affected. For instance, mutations near the beginning of the coding sequence generally result in more severe phenotypic outcomes compared to those closer to the end.
- Therapeutic Implications: Understanding the mechanisms underlying nonsense mutations is crucial for developing targeted therapies. For example, strategies such as nonsense codon read-through drugs aim to bypass premature stop codons and restore protein function in patients with genetic diseases caused by nonsense mutations.
Outcomes of a Nonsense Mutation
- Deleterious Outcomes
- Description: Nonsense mutations often result in the early termination of protein synthesis, producing truncated proteins that lack essential functional domains. Such mutations can severely impair the protein’s ability to perform its intended function, leading to significant biological consequences.
- Impact: These mutations frequently contribute to a decline in reproductive fitness and overall organismal health. This is due to the critical roles that these proteins play in various physiological processes.
- Example: A classic example is the nonsense mutation in the CFTR gene, which leads to cystic fibrosis. The premature stop codon results in a truncated CFTR protein that cannot regulate ion transport effectively, causing severe respiratory and digestive issues.
- Neutral Outcomes
- Description: Some nonsense mutations occur in regions of a gene where their effects are minimal or nonexistent. These mutations do not significantly alter the protein’s function or the organism’s phenotype.
- Impact: Organisms carrying such mutations generally experience no detectable benefit or detriment. These mutations do not influence the protein’s ability to interact with other molecules or perform its biological role.
- Example: A nonsense mutation occurring in the coding region just before the final amino acid of a protein may not significantly affect the protein’s function. As a result, the mutation has a neutral impact on the organism.
- Beneficial Outcomes
- Description: Although rare, some nonsense mutations can provide advantageous effects. These mutations can sometimes lead to proteins with modified functions that improve the organism’s survival or reproductive success in specific environments.
- Impact: Beneficial nonsense mutations enhance the organism’s fitness by conferring resistance to environmental stressors or improving metabolic processes.
- Example: A hypothetical example could be a nonsense mutation in a channel protein that alters its function, thereby preventing the entry of harmful toxins into the cell. This could offer a selective advantage in environments where exposure to toxins is prevalent.
Mechanism of nonsense mutation
Here is a detailed explanation of the mechanisms involved in nonsense mutations:
- Tautomerism: One of the primary mechanisms leading to nonsense mutations is tautomerism. Nucleotides exist in two tautomeric forms: the more stable keto form and the less common enol form. The keto form typically forms stable hydrogen bonds with complementary nucleotides, facilitating accurate base pairing during DNA replication. However, tautomeric shifts can occur, where the keto form converts to the enol form. This conversion alters the hydrogen bonding pattern, potentially causing base pair mismatches and the subsequent introduction of a stop codon.
- Ionization: Ionizing agents, such as radiation, can induce changes in nucleotide structures, leading to mutations. Ionization can alter the chemical properties of nucleotides, increasing the likelihood of base mispairing or improper bonding during DNA replication. This can result in the formation of stop codons, contributing to nonsense mutations.
- Mutagenesis by Chemical Agents: Chemical mutagens can introduce nucleotide substitutions directly into the DNA sequence. These agents may modify the nucleotide bases, leading to the substitution of a sense codon with a stop codon. For example, specific chemicals might cause the conversion of codons that normally code for amino acids into premature stop codons.
- Enzymatic Digestion: During DNA replication or repair processes, nucleases and other enzymes may inadvertently remove nucleotides from the DNA sequence. The loss of nucleotides can lead to frameshift mutations, which may eventually result in the introduction of a premature stop codon due to altered reading frames or codon shifts.
- Splicing Errors: In some instances, nonsense mutations can be the result of errors during RNA splicing. If the splicing machinery incorrectly processes the pre-mRNA, it can introduce or skip exons containing premature stop codons, affecting the final mRNA sequence and resulting in a truncated protein.
- Multiple Codon Changes: A single nucleotide change can create one of the three stop codons (UAG, UAA, or UGA). Because these stop codons are the result of specific nucleotide substitutions, multiple single-nucleotide changes can produce the same stop codon, leading to a premature termination of protein synthesis.
- Genetic Variation and Mutagenesis: Genetic variations and spontaneous mutations can also contribute to the formation of nonsense mutations. These include natural errors in DNA replication and the presence of genetic predispositions that increase susceptibility to such mutations.
- De Novo Mutations: Nonsense mutations can arise spontaneously in the germline cells, termed de novo mutations. These mutations occur without prior exposure to mutagens and result from random errors in DNA replication or repair mechanisms.
- Functional Implications: The impact of nonsense mutations on protein function depends on their position within the coding sequence. Mutations occurring early in the coding sequence generally result in more severe phenotypic consequences compared to those occurring later.
- Disease Associations: Nonsense mutations are associated with various genetic disorders. For example, these mutations can cause diseases such as Duchenne muscular dystrophy and cystic fibrosis, where the truncated proteins lead to loss of function and severe clinical manifestations.
Genetic diseases or disorders Associated with Nonsense Mutations
Nonsense mutations can lead to a variety of genetic disorders by truncating proteins essential for normal cellular function. These mutations introduce premature stop codons in the coding sequence of a gene, leading to incomplete and non-functional protein products. The impact of nonsense mutations can vary widely, but they are often associated with serious and potentially life-threatening conditions.
- Cystic Fibrosis
- Description: Cystic fibrosis (CF) is a genetic disorder caused by nonsense mutations in the CFTR gene, which encodes the cystic fibrosis transmembrane conductance regulator protein. The G542X mutation is one such example where a glycine codon is replaced by a stop codon, leading to a non-functional CFTR protein.
- Impact: This results in defective ion channel function and impaired chloride transport across epithelial cell membranes. The accumulation of thick mucus in various organs, particularly the lungs, leads to severe respiratory and digestive problems.
- Example: The G542X mutation in CFTR is a well-known cause of CF, leading to the complete absence of functional CFTR protein.
- Beta-Thalassemia
- Description: Beta-thalassemia is a blood disorder resulting from nonsense mutations in the beta-globin gene. This gene encodes one of the subunits of hemoglobin, the oxygen-carrying molecule in red blood cells. The nonsense mutations result in truncated beta-globin proteins.
- Impact: The production of abnormal hemoglobin leads to reduced oxygen transport and anemia. This condition results in fatigue, weakness, and other complications related to insufficient oxygen delivery to tissues.
- Example: Various nonsense mutations in the beta-globin gene can lead to beta-thalassemia, significantly affecting oxygen transport and causing anemia.
- Hurler Syndrome
- Description: Hurler syndrome is caused by nonsense mutations in the IDUA gene, which codes for the alpha-L-iduronidase enzyme. This enzyme is crucial for the breakdown of glycosaminoglycans (GAGs).
- Impact: A deficiency in alpha-L-iduronidase leads to the accumulation of GAGs, causing progressive damage to tissues and organs. The symptoms include developmental delays, skeletal abnormalities, and cognitive impairment. If untreated, Hurler syndrome can be fatal in early childhood.
- Example: The nonsense mutation in the IDUA gene results in the absence of functional enzyme activity, leading to the characteristic features of Hurler syndrome.
- Duchenne Muscular Dystrophy (DMD)
- Description: Duchenne muscular dystrophy is a severe form of muscular dystrophy caused by nonsense mutations in the DMD gene, which encodes the dystrophin protein. The mutation introduces premature stop codons, resulting in a truncated dystrophin protein.
- Impact: The absence of functional dystrophin compromises muscle cell integrity, leading to progressive muscle weakness and degeneration. This condition predominantly affects males and often leads to severe mobility impairment and life-threatening complications.
- Example: Nonsense mutations in the DMD gene are a major cause of Duchenne muscular dystrophy, and treatments like ataluren aim to read-through the premature stop codons to produce functional dystrophin.
Examples of nonsense mutation
- Duchenne Muscular Dystrophy (DMD): One notable example of a nonsense mutation is observed in Duchenne muscular dystrophy (DMD). In this genetic disorder, nonsense mutations in the dystrophin gene can introduce premature stop codons. These mutations lead to truncated dystrophin proteins that are nonfunctional, contributing to the progressive muscle degeneration characteristic of the disease.
- Cystic Fibrosis (CF): Another example is found in cystic fibrosis, where nonsense mutations in the CFTR gene can result in premature stop codons. These mutations disrupt the synthesis of the CFTR protein, which is crucial for regulating ion channels in epithelial cells. The lack of functional CFTR protein leads to the accumulation of thick mucus in various organs, including the lungs and pancreas.
- Spinal Muscular Atrophy (SMA): Nonsense mutations in the survival motor neuron 1 (SMN1) gene are implicated in spinal muscular atrophy. These mutations create premature stop codons that produce truncated SMN proteins. The reduced levels of functional SMN protein affect motor neuron survival, leading to muscle weakness and atrophy.
- Osteogenesis Imperfecta (OI): In osteogenesis imperfecta, certain nonsense mutations in the COL1A1 or COL1A2 genes introduce early stop codons. These mutations disrupt the production of type I collagen, a critical component of connective tissue. The resulting collagen is defective, leading to fragile bones and other connective tissue abnormalities.
- Hemophilia B: Hemophilia B, a bleeding disorder, can be caused by nonsense mutations in the F9 gene, which encodes for coagulation factor IX. These mutations result in truncated factor IX proteins, impairing blood clotting and leading to excessive bleeding tendencies.
- Beta-Thalassemia: Nonsense mutations in the HBB gene, responsible for encoding beta-globin, can cause beta-thalassemia. Such mutations introduce premature stop codons that lead to reduced production of beta-globin chains. This imbalance results in ineffective red blood cell production and anemia.
- Tay-Sachs Disease: In Tay-Sachs disease, nonsense mutations in the HEXA gene can create premature stop codons. The HEXA gene encodes the alpha subunit of hexosaminidase A, an enzyme essential for breaking down GM2 gangliosides. Deficient enzyme activity leads to the accumulation of toxic substances in nerve cells, causing severe neurological damage.
- Retinitis Pigmentosa: Nonsense mutations in the RHO gene, which encodes rhodopsin, can lead to retinitis pigmentosa. These mutations introduce stop codons that truncate the rhodopsin protein, disrupting photoreceptor function in the retina and leading to progressive vision loss.
- Ataxia Telangiectasia: Nonsense mutations in the ATM gene, which encodes a protein involved in DNA repair, are linked to ataxia telangiectasia. These mutations result in truncated ATM proteins, compromising the cell’s ability to repair DNA damage and leading to neurodegeneration and immune system deficiencies.
- Hereditary Breast and Ovarian Cancer: Nonsense mutations in the BRCA1 or BRCA2 genes can lead to hereditary breast and ovarian cancer. These mutations introduce premature stop codons that produce truncated proteins involved in DNA repair. The loss of functional BRCA proteins impairs genomic stability and increases cancer risk.
Applications of nonsense mutation
- Therapeutic Approaches: Nonsense mutations can be strategically used in gene therapy to combat cancer. By introducing nonsense mutations into genes associated with uncontrolled cell proliferation, researchers can induce premature termination of translation, leading to truncated and nonfunctional proteins. This can help in preventing the production of oncogenic proteins that drive cancerous growth. Therefore, targeting specific genes with nonsense mutations might offer a method to halt or slow down tumor progression.
- Enhancing Organism Fitness: In evolutionary biology, nonsense mutations can sometimes lead to advantageous traits that improve an organism’s fitness in a particular environment. Such mutations may alter protein functions in ways that confer survival benefits. For instance, a nonsense mutation might disrupt a protein that inhibits an adaptive trait, allowing the organism to better cope with environmental challenges.
- Disease Research: Nonsense mutations are implicated in approximately 5-15% of genetic diseases. Understanding these mutations is crucial for diagnosing and developing treatments for conditions such as Duchenne muscular dystrophy, cystic fibrosis, and various cancers. By studying the specific nonsense mutations responsible for these diseases, researchers can gain insights into disease mechanisms and potential therapeutic targets.
- Protein Stability and Function Studies: In laboratory settings, nonsense mutations are employed to investigate protein stability and function. By introducing nonsense mutations into specific genes, scientists can create truncated versions of proteins. This approach allows researchers to analyze how the loss of functional domains affects protein behavior and interactions, providing valuable information about protein structure-function relationships.
- Synthetic Biology: Nonsense mutations are utilized in synthetic biology to design genetic circuits and biosensors. By incorporating stop codons into synthetic constructs, researchers can control the production of proteins and regulate gene expression. This technique is useful for creating precise genetic tools and optimizing synthetic pathways for industrial applications.
- Understanding Genetic Pathways: Nonsense mutations can help elucidate genetic pathways and networks by disrupting specific genes. The resulting phenotypic changes can reveal the role of the mutated gene within broader biological processes. This information contributes to a deeper understanding of genetic interactions and regulatory mechanisms.
- Gene Knockout Models: Nonsense mutations are used to create gene knockout models in experimental organisms. These models allow scientists to study the effects of gene loss on development, physiology, and disease. By observing the consequences of premature stop codons, researchers can better understand the functions of essential genes.
- Drug Development: In the field of drug development, nonsense mutations are used to identify potential drug targets and mechanisms of action. Understanding how drugs affect proteins with nonsense mutations can guide the development of therapies aimed at correcting or mitigating the effects of these mutations.
- Genetic Screening: Nonsense mutations are important in genetic screening programs, especially for inherited disorders. Identifying nonsense mutations in individuals can provide diagnostic information, help in assessing disease risk, and guide personalized treatment strategies.
- Evolutionary Studies: Nonsense mutations contribute to studies on genetic evolution and diversity. By examining how these mutations influence fitness and adaptation, researchers can gain insights into evolutionary processes and the role of genetic variations in shaping species.
Facts
- Did you know that nonsense mutations introduce premature stop codons into the genetic sequence, leading to truncated and often nonfunctional proteins?
- Have you heard that nonsense mutations can occur due to various causes, including nucleotide substitutions and errors during DNA replication?
- Are you aware that nonsense mutations are implicated in several genetic diseases, such as cystic fibrosis, Duchenne muscular dystrophy, and beta-thalassemia, due to their disruptive effects on protein synthesis?
- Can you believe that the severity of a genetic disease caused by a nonsense mutation can vary depending on the location of the stop codon within the gene?
- Did you know that nonsense mutations often result in nonsense-mediated mRNA decay (NMD), a cellular process that degrades mRNA transcripts containing premature stop codons?
- Have you heard that the premature stop codons introduced by nonsense mutations can lead to the complete loss of protein function or the production of a malfunctioning protein?
- Are you aware that nonsense mutations can sometimes be used intentionally in research to study the function of specific proteins and their role in cellular processes?
- Can you believe that the introduction of nonsense mutations in experimental models can help scientists understand the genetic basis of various diseases and develop targeted therapies?
- Did you know that nonsense mutations in the CFTR gene cause cystic fibrosis by leading to a nonfunctional CFTR protein, which disrupts ion transport and causes severe respiratory symptoms?
- Have you heard that in Duchenne muscular dystrophy, nonsense mutations in the DMD gene result in truncated dystrophin proteins, leading to progressive muscle weakness and degeneration?
- Are you aware that beta-thalassemia is often caused by nonsense mutations in the beta-globin gene, resulting in reduced production of functional hemoglobin and anemia?
- Can you believe that some nonsense mutations lead to genetic conditions such as Hurler syndrome by disrupting the production of enzymes essential for breaking down complex sugars?
- Did you know that certain therapies, such as ataluren, are designed to bypass premature stop codons caused by nonsense mutations, potentially restoring protein function in genetic disorders?
- Have you heard that the effects of nonsense mutations can sometimes be mitigated by using gene editing technologies to correct the mutations and restore normal protein function?
- Are you aware that the study of nonsense mutations and their consequences provides valuable insights into gene function, protein synthesis, and the development of genetic therapies?
- Mort, M., Ivanov, D., Cooper, D., & Chuzhanova, N. (2008). A meta‐analysis of nonsense mutations causing human genetic disease. Human Mutation, 29. https://doi.org/10.1002/humu.20763.
- Arboleda, V., Lee, H., Dorrani, N., Zadeh, N., Willis, M., Macmurdo, C., Manning, M., Kwan, A., Hudgins, L., Barthélémy, F., Miceli, M., Quintero-Rivera, F., Kantarci, S., Strom, S., Deignan, J., Grody, W., Vilain, E., & Nelson, S. (2015). De novo nonsense mutations in KAT6A, a lysine acetyl-transferase gene, cause a syndrome including microcephaly and global developmental delay.. American journal of human genetics, 96 3, 498-506 . https://doi.org/10.1016/j.ajhg.2015.01.017.
- Dietz, H., Valle, D., Francomano, C., Kendzior, R., Pyeritz, R., & Cutting, G. (1993). The skipping of constitutive exons in vivo induced by nonsense mutations. Science, 259, 680 – 683. https://doi.org/10.1126/SCIENCE.8430317.
- Benhabiles, H., Jia, J., & Lejeune, F. (2016). General Aspects Related to Nonsense Mutations. , 1-76. https://doi.org/10.1016/B978-0-12-804468-1.00001-4.
- Zarraga, I., Zhang, L., Stump, M., Gong, Q., Vincent, G., & Zhou, Z. (2011). Nonsense-mediated mRNA decay caused by a frameshift mutation in a large kindred of type 2 long QT syndrome.. Heart rhythm, 8 8, 1200-6 . https://doi.org/10.1016/j.hrthm.2011.03.039.
- Stella, A., Wagner, A., Shito, K., Lipkin, S., Watson, P., Guanti, G., Lynch, H., Fodde, R., & Liu, B. (2001). A nonsense mutation in MLH1 causes exon skipping in three unrelated HNPCC families.. Cancer research, 61 19, 7020-4 .
- Torella, A., Zanobio, M., Zeuli, R., Blanco, F., Savarese, M., Giugliano, T., Garofalo, A., Piluso, G., Politano, L., & Nigro, V. (2020). The position of nonsense mutations can predict the phenotype severity: A survey on the DMD gene. PLoS ONE, 15. https://doi.org/10.1371/journal.pone.0237803.
- https://journals.plos.org/plosone/article?id=10.1371/journal.pone.0237803
- https://biologydictionary.net/nonsense-mutation/#outcomes-of-a-nonsense-mutation
- https://www.geeksforgeeks.org/difference-between-missense-and-nonsense-mutation/
- https://www.technologynetworks.com/genomics/articles/missense-nonsense-and-frameshift-mutations-a-genetic-guide-329274
- https://www.biologyonline.com/dictionary/nonsense-mutation#Nonsense_Mutation_Example
- https://en.wikipedia.org/wiki/Nonsense_mutation