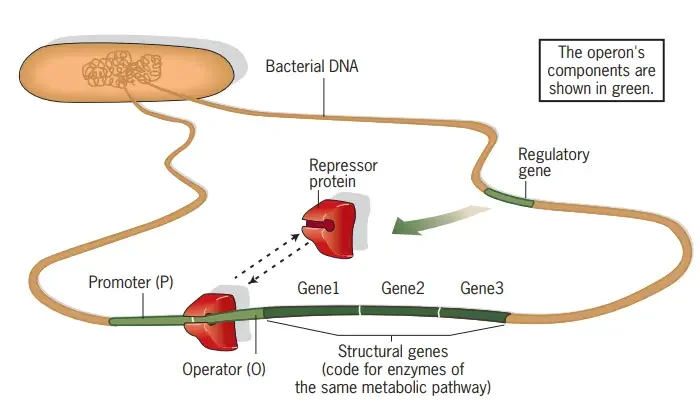
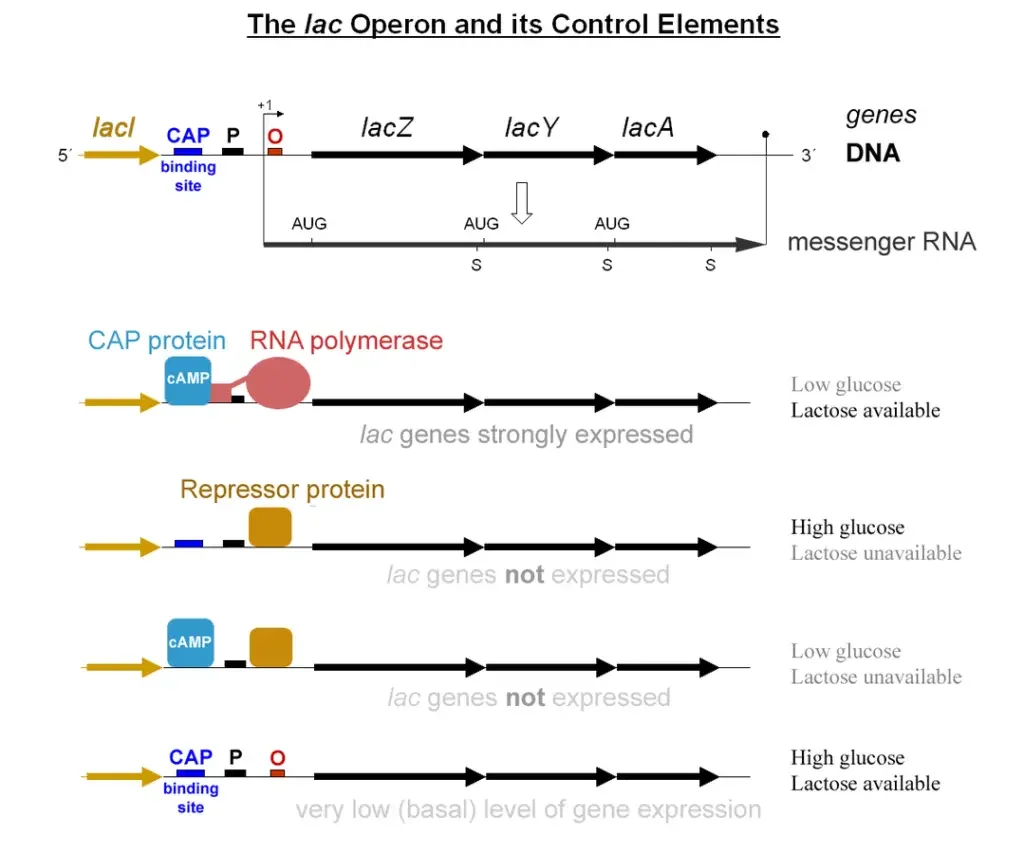
What is lac Operon?
- The lac operon, also known as the lactose operon, is a genetic regulatory system found in Escherichia coli (E. coli) and other enteric bacteria. It plays a crucial role in the transport and metabolism of lactose, a disaccharide sugar. While glucose is the preferred carbon source for most bacteria, the lac operon enables efficient digestion of lactose when glucose is not available.
- The lac operon consists of a cluster of genes that work together to facilitate lactose metabolism. When lactose is needed as a sugar source, the three genes within the lac operon are expressed and their corresponding proteins are synthesized. These genes are lacZ, lacY, and lacA. The lacZ gene encodes β-galactosidase, an enzyme responsible for breaking down lactose into glucose and galactose. The lacY gene produces Beta-galactoside permease, a membrane protein that aids in the transport of lactose into the bacterial cell. Finally, the lacA gene encodes β-galactoside transacetylase, which participates in the acetylation of certain β-galactosides.
- However, it would be energetically wasteful for the cell to produce these enzymes when lactose is unavailable or when a more favorable energy source like glucose is present. To prevent unnecessary enzyme production, the lac operon employs a two-part control mechanism.
- In the absence of lactose, a regulatory protein called the lac repressor (encoded by the lacI gene) binds to the operator region of the lac operon. This binding prevents the expression of the lac genes, effectively halting the production of the enzymes required for lactose metabolism. The lac repressor is always present in the cell but remains inactive unless it binds to a small co-inducer molecule.
- The lac operon’s second control mechanism involves the presence of glucose. When glucose is available, the catabolite activator protein (CAP) remains inactive. CAP is necessary for the full activation of the lac operon. Additionally, the protein EIIAGlc inhibits lactose permease, preventing the transport of lactose into the cell.
- This dual control mechanism ensures that the lac operon is activated only when lactose is present and glucose is scarce. It allows the sequential utilization of glucose and lactose in two distinct growth phases known as diauxie. The lac operon’s regulation and function were extensively studied by François Jacob and Jacques Monod, and their work on the lac operon led to their recognition with the Nobel Prize in Physiology in 1965.
- Overall, the lac operon serves as a paradigmatic example of gene regulation in prokaryotic organisms. Its study has provided valuable insights into the mechanisms of gene expression and cellular adaptation to different nutrient conditions.
Jacques Monod (1910–1976) began studying bacterial growth and regulation in the late 1930s. He selected E. coli as a model bacteria and eventually concentrated on genes involved in E. coli’s growth on lactose. Francois Jacob joined him in his studies roughly 15 years later (1920–2013). As a result of their research, the lactose (lac) operon of E. coli is likely the most thoroughly investigated negative control system.
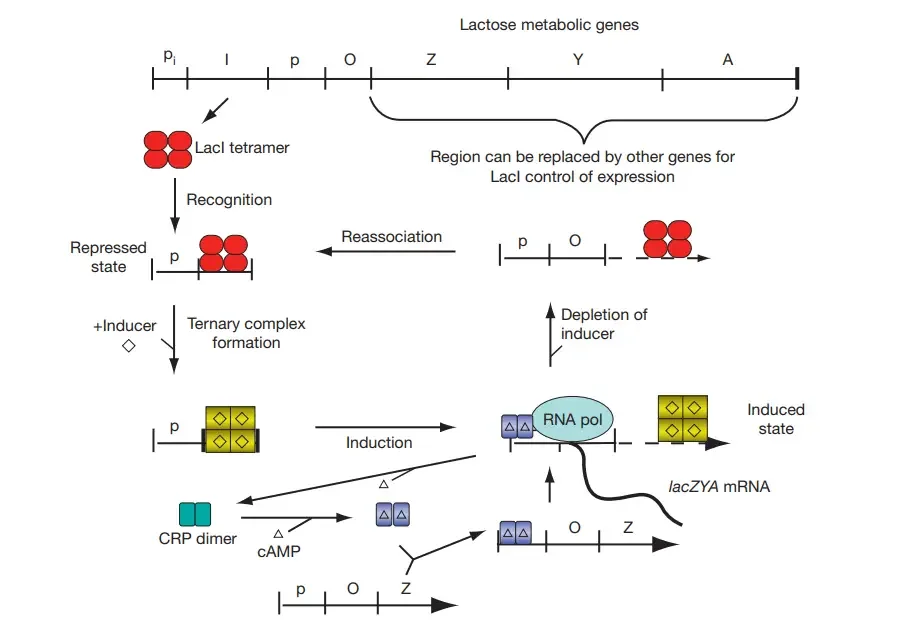
About Above Image: The upper horizontal bar represents regions of the lac operon DNA: the promoter (pi) for the tetrameric lactose repressor (lacI gene, ‘I’), the promoter (p) for the metabolic enzymes, the primary operator sequence LacO (‘O’, LacI binding site), and the co-transcribed metabolic enzyme genes (lacZYA, ‘Z,’ ‘Y,’ and ‘A’). LacI tetramer (red ovals) binding to operator, LacI association with inducer (diamond) to form LacI-inducer complex (diamond within yellow squares), LacI dissociation from operator (induction), RNA polymerase (‘RNA pol’) binding and transcription, and subsequent reassociating of LacI with LacO when sugar levels decrease. CRP interaction with cAMP (triangle) to produce the CRP–cAMP complex (triangles inside purple rectangles) and CRP–cAMP binding within the promoter region to aid RNA polymerase binding and transcription are depicted at the bottom of the figure. When glucose levels rise and cAMP levels fall, the CRP dissociates from the promoter. Changes in color/shape of protein symbols signify conformational alterations in response to ligand binding (inducer or cAMP). Substitution of lac metabolic genes with other (prokaryotic or eukaryotic) protein-coding sequences enables excellent control of specific protein expression utilising inducer/cAMP levels.
Lac operon definition
The lac operon is a genetic regulatory system found in bacteria, such as E. coli, that controls the transport and metabolism of lactose. It consists of a cluster of genes that are expressed and produce enzymes when lactose is available, while remaining inactive in the absence of lactose or when a more favorable energy source, like glucose, is present.
Features of Lac Operon
The lac operon, a gene cluster present in certain bacterial species, orchestrates the metabolism of lactose, a disaccharide prevalent in milk. This operon exemplifies the intricate regulatory mechanisms that bacteria employ to adapt to varying environmental conditions. Below are the salient features of the lac operon:
- Structural Genes: The lac operon encompasses three primary structural genes:
- lacZ: Encodes for β-galactosidase, an enzyme that facilitates the hydrolysis of lactose into glucose and galactose.
- lacY: Produces lactose permease, a protein responsible for the transport of lactose across the bacterial cell membrane.
- lacA: Transcribes for transacetylase, an enzyme with a less defined role in lactose metabolism.
- Regulatory Gene: Apart from the structural genes, the operon houses a regulatory gene, lacI, which synthesizes the lac repressor protein. This repressor binds to the operator segment of the operon, inhibiting the transcription of the structural genes in the absence of lactose.
- Inducibility: The lac operon operates on an inducible system. In the milieu of lactose, the sugar molecule interacts with the lac repressor, inducing a structural alteration that dislodges the repressor from the operator, thereby initiating transcription.
- Catabolite Repression: Despite the presence of lactose, the transcription of the lac operon can be suppressed by glucose, a more energetically favorable carbon source for bacteria. This phenomenon, termed catabolite repression, ensures that bacteria prioritize glucose metabolism over lactose.
- Dual Regulation: The lac operon’s transcription is modulated by both positive and negative feedback mechanisms. The catabolite activator protein (CAP) exemplifies positive regulation. In environments with diminished glucose levels, CAP binds upstream of the promoter, amplifying transcription rates.
- Attenuation: Beyond the primary regulatory mechanisms, the lac operon employs attenuation for refined control over gene expression. This process hinges on the mRNA’s capability to adopt diverse stem-loop configurations, influencing the progression of transcription elongation.
- Mutational Impacts: Perturbations in the lac operon, stemming from mutations, can drastically alter lactose metabolic regulation. Such changes can influence the bacterial survival rate and adaptability across diverse habitats, underscoring the operon’s evolutionary significance.
Structure of the lac operon
The components of a lac operon are Structural genes and Regulatory DNA sequences.
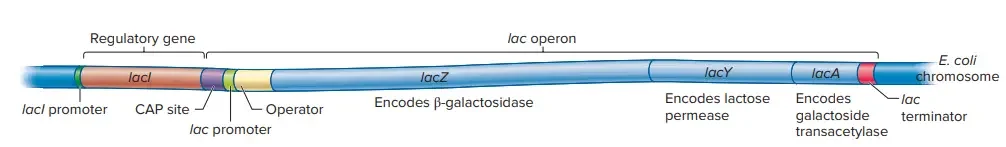
Structural genes of lac operon
The lac operon consists of three structural genes: lacZ, lacY, and lacA. These genes are transcribed together as a single polycistronic mRNA from a common promoter.
- lacZ: This gene codes for the enzyme β-galactosidase, which is a tetramer with a molecular weight of approximately 500 kD. β-galactosidase plays a crucial role in lactose metabolism by breaking down β-galactosides, including lactose, into their monosaccharide components. For example, lactose is hydrolyzed into glucose and galactose, which can be further metabolized through glycolysis.
- lacY: This gene encodes the β-galactoside permease, which is a membrane-bound protein with a molecular weight of about 30 kD. β-galactoside permease facilitates the transport of β-galactosides, such as lactose, into the bacterial cell. It is responsible for the uptake of lactose from the environment.
- lacA: This gene codes for β-galactoside transacetylase, although its precise role within the lac operon is not fully understood. β-galactoside transacetylase transfers an acetyl group from acetyl-CoA to β-galactosides. However, its specific function in lactose metabolism remains unclear.
These three structural genes, lacZ, lacY, and lacA, are located adjacent to each other within the lac operon. Together with the promoter, operator, and regulatory elements, they form a functional unit known as an operon. The lac operon provides the necessary genes and enzymes for the uptake and metabolism of lactose and other β-galactosides in bacteria like E. coli.
Regulatory genes of lac operon
The lac operon consists of several regulatory genes that control its activity:
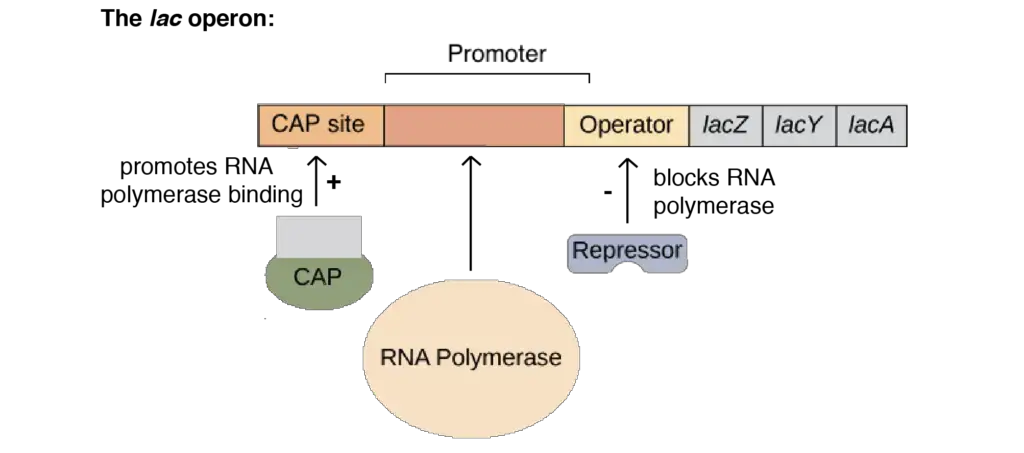
- Promoter: The promoter region is the binding site for RNA polymerase, the enzyme responsible for initiating transcription. It is located upstream of the lac operon and facilitates the binding of RNA polymerase to initiate the transcription of the structural genes.
- Operator: The operator region is a negative regulatory site situated between the promoter and the structural genes. It overlaps with the promoter region. The lac repressor protein binds to the operator, preventing RNA polymerase from transcribing the structural genes. The operator acts as a switch, determining whether transcription should occur or not.
- Lac I (Repressor) Gene: The Lac I gene codes for the lac operon repressor protein. It is located adjacent to the promoter region of the lac operon and has its own promoter and terminator. The repressor protein is a tetramer composed of identical subunits with a molecular weight of 38 kD each. The repressor is continuously synthesized since its gene is always transcribed. The repressor protein can bind to the operator, thereby repressing (turning off) the lac operon by blocking RNA polymerase binding.
- Catabolite Activator Protein (CAP) Binding Site: The CAP binding site is a positive regulatory site found just upstream of the lac operon promoter. The catabolite activator protein (CAP) binds to this site. CAP is a dimeric protein that can bind to cAMP (cyclic adenosine monophosphate) and DNA. When cAMP binds to CAP, its affinity for DNA increases. CAP bound to DNA promotes transcription by enhancing the binding of RNA polymerase to the promoter region, leading to increased gene expression of the lac operon.
These regulatory genes, including the promoter, operator, Lac I repressor gene, and CAP binding site, work together to control the activity of the lac operon, allowing the cell to respond to the presence or absence of lactose and other regulatory signals.
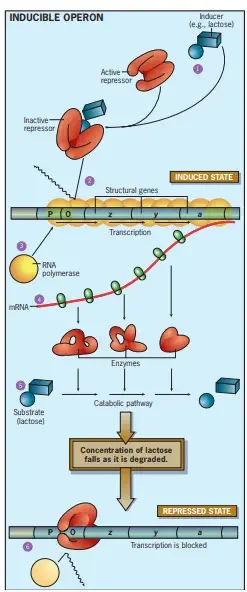
The lac repressor
- The lac repressor is a protein that suppresses (represses) lac operon transcription. This is accomplished by binding to the operator, which partially overlaps the promoter.
- When bound, lac repressor impedes RNA polymerase and prevents transcription of the operon.
- The gene lacI, which encodes the lac repressor, is under the control of its own promoter.
- The lacI gene is located in close proximity to the lac operon, however it is not a component of the operon and is expressed independently. lacI is continuously transcribed, therefore its protein product, lac repressor, is consistently present.
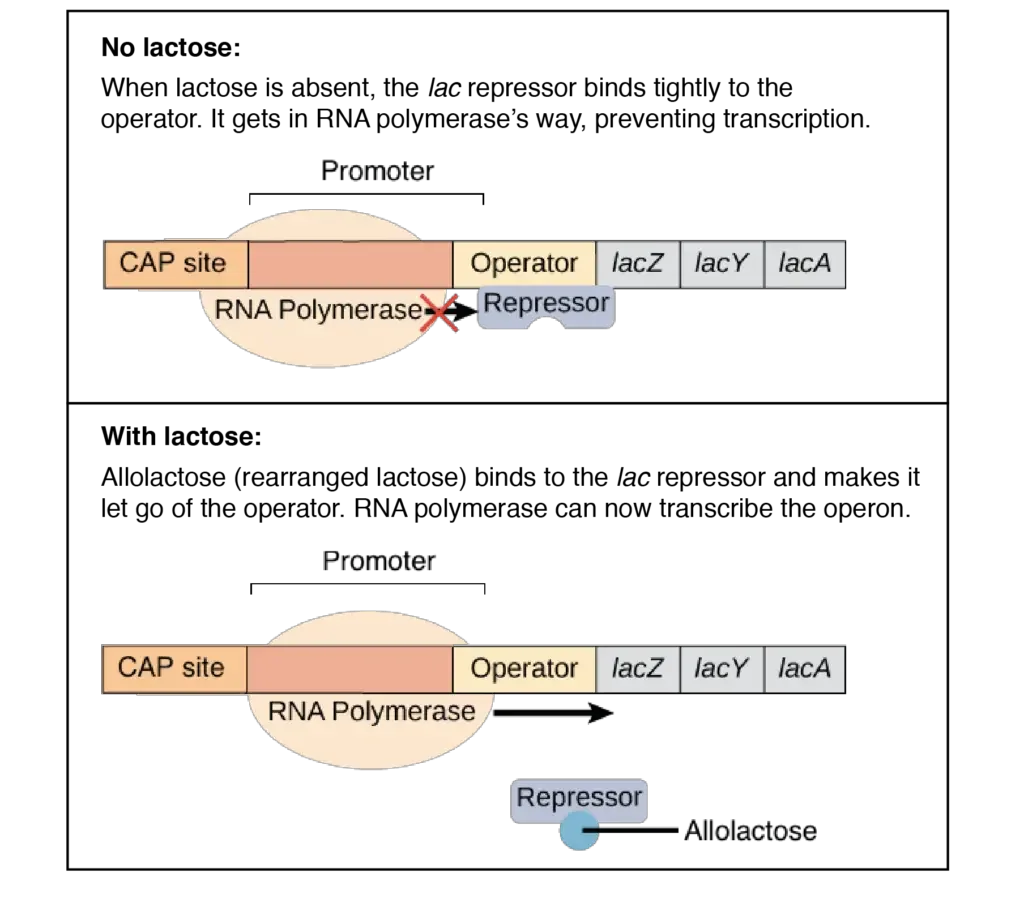
- When lactose is unavailable, the lac repressor binds strongly to the operator, inhibiting RNA polymerase transcription.
- In the presence of lactose, the lac repressor loses its capacity to bind DNA. It detaches itself from the operator, allowing RNA polymerase to transcribe the operon.
- This alteration in the lac repressor is caused by the lactose isomer allolactose.
- When lactose is present, certain molecules within the cell will be transformed to allolactose. Allolactose binds to lac repressor and causes it to shift form, preventing it from binding DNA.
- Allolactose is an example of an inducer, a tiny chemical that stimulates gene or operon expression.
- The lac operon is an inducible operon because it is normally turned off (repressed), but can be activated in the presence of allolactose.
LacI Function – Mechanism
- In the absence of lactose, LacI inhibits the synthesis of mRNA encoding proteins expressed by the lac operon.
- Although transcription is not eliminated entirely, lacZYA mRNA is transcribed at extremely low quantities. LacI protein binds with high specificity and affinity to the lac operator DNA sequence to perform this activity.
- Consequently, transcription is blocked by a variety of methods. Since the lac operator (LacO) overlaps the promoter, binding of LacI competes directly with RNA polymerase for binding to this region.
- LacI can also inhibit transcription start and/or mRNA elongation to effectively repress transcription.
- When lactose is available as a carbon source, lactose is transported into the bacterium by low quantities of the metabolic protein LacY.
- Next, residual LacZ converts lactose into glucose and galactose, which provides the bacterium with energy.
- Notably, this catalytic mechanism also produces low quantities of allolactose by rearranging the b-1,4 connection between glucose and galactose to a b-1,6 linkage.
- Allolactose binds to LacI and induces a conformational shift in the repressor protein, causing the operator DNA sequence to be released (induction).
- Thus, RNA polymerase is liberated to produce multiple copies of mRNA encoding lac metabolic proteins.
- Utilizing an environmental opportunity, these metabolic proteins, when translated into proteins, enable the bacterium to transport and digest huge volumes of lactose as its carbon energy source.
- Jacob and Monod discovered that a range of non-natural galactoside sugars (such as IPTG) can stimulate LacI and ease transcription inhibition of lacZYA as a consequence of their research. This discovery has had a significant impact on the biotechnology applications of this regulatory system.
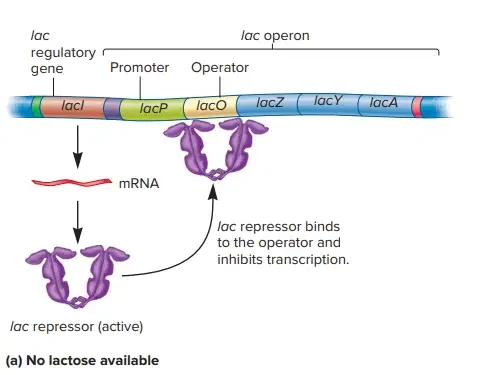
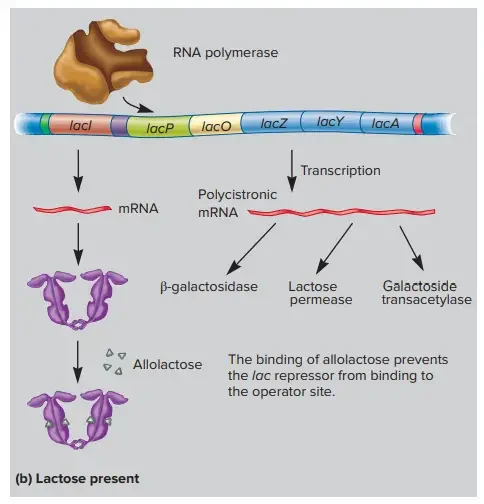
(a) The lac repressor is active and binds the operator when allolactose is not present. Repressor binding to the operator inhibits transcription. (b) When lactose is available, some of it is converted to allolactose by β-galactosidase. When sufficient amounts of allolactose are present, it binds and inactivates the lac repressor. The repressor leaves the operator and RNA polymerase is free to initiate transcription.
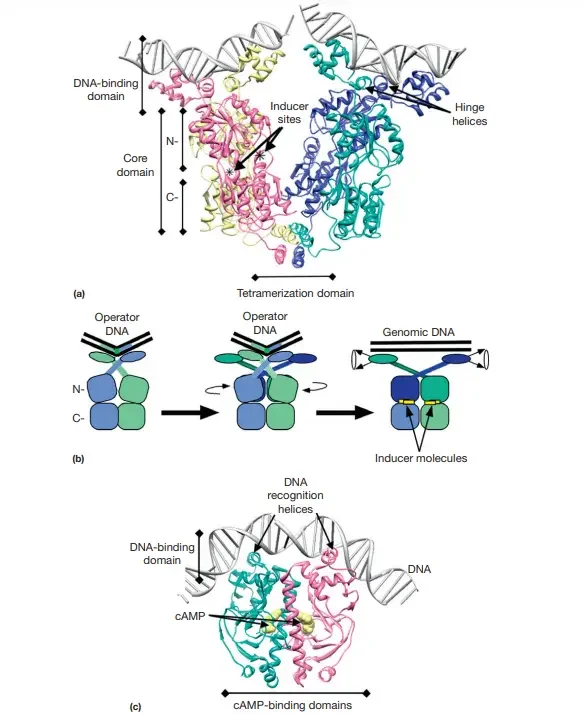
Structural Characteristics of LacI
The atomic-level process by which the LacI protein executes its tasks has been illuminated by LacI’s structural specifics.
- LacI is a multimeric, multidomain protein, a configuration that is now recognised as being typical of genetic regulatory proteins.
- Four identical polypeptide chains comprise LacI. Each polypeptide folds into different functional areas, and four monomers combine to produce tetrameric LacI.
- Folded protein domains can be separated from the intact protein while retaining functions that are remarkably similar to those of the full protein.
- Proteolysis can be used to isolate the N-terminal domain of a LacI monomer, with or without portions of the hinge-helix sequence.
- An isolated N-terminal domain can bind with low affinity to one-half of LacO DNA. This protein fragment binds to full-length LacO with particularly high affinity if two of these domains and the hinge-helix sequences are connected by a disulfide bond (but is not inducible by galactoside sugars). (Note that the LacO site is strongly but imperfectly symmetric in sequence, allowing for the presence of two identical DNA-binding domains.)
- Only binding with high affinity by LacI inhibits mRNA transcription. Consequently, the LacI dimer is the functional unit, and a LacI tetramer has two DNA-binding sites.
- Each N-terminal domain of the LacI–LacO complex connects with DNA bases in the major groove, while the hinge helices insert into the minor groove to induce a DNA bend.
- LacI’s core domain is the second result of proteolysis. This segment is isolated as a tetramer, each monomer containing two subdomains that flank the sugar-binding site.
- Consequently, each monomer can bind an inducer molecule, resulting in four sugar-binding sites per tetramer. The N- and C-subdomains of the core domain monomer are coupled by numerous strands and cannot be dissociated by proteolysis.
- When the inducer-binding site is occupied in full-length LacI, the DNA-binding domain undergoes structural modifications in the N-subdomain.
- Because of these changes, the operator-specific binding affinity decreases, and the repressor protein that is bound to the inducer separates from the operator DNA site and moves to other parts of the genomic DNA.
- The C-terminal tetramerization domain of LacI joins two dimers by means of an antiparallel fourhelix bundle. When this domain is deleted, the resultant LacI dimer is still capable of repressing transcription.
Catabolite activator protein (CAP)
- Catabolite activator protein, CAP (also known as cAMP receptor protein, CRP) is essential for high levels of lac operon transcription.
- It forms a CRP–cAMP complex in association with 3’5′ cyclic AMP.
- CRP–cAMP binds to the lac promoter and enhances RNA polymerase binding, hence driving transcription of the lac operon.
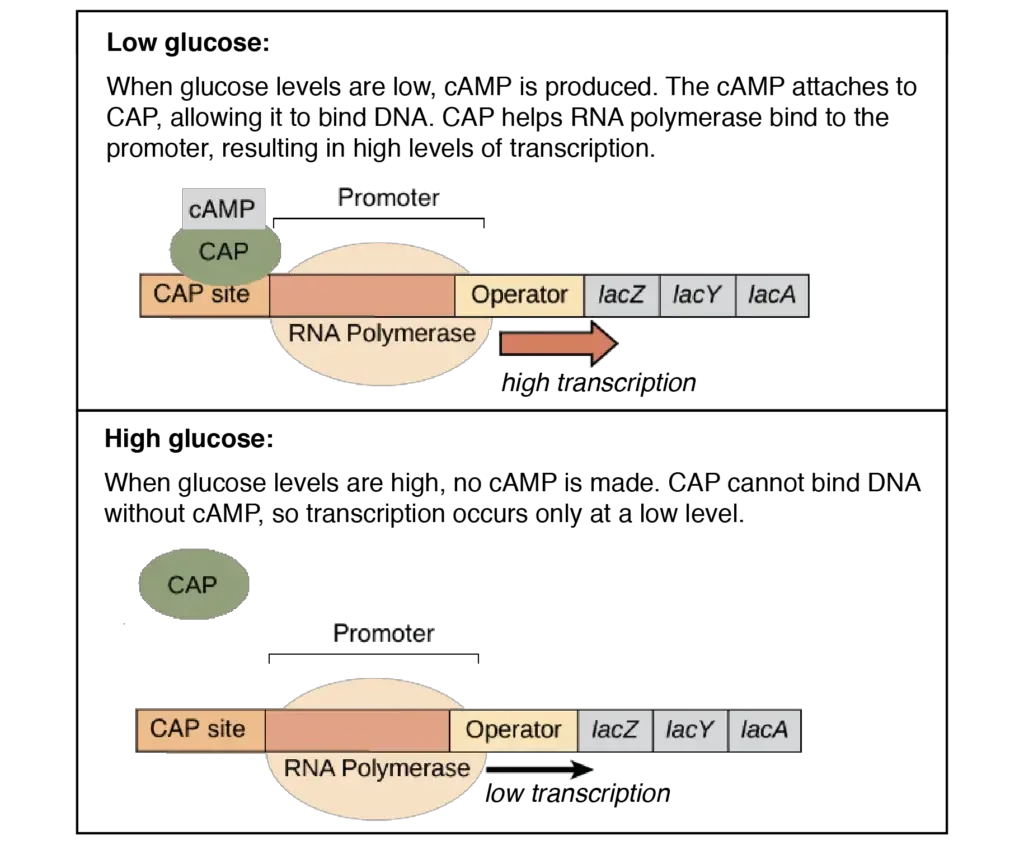
- When glucose is available, intracellular cAMP levels decrease, CRP cannot bind to the lac promoter on its own, and the lac operon is only faintly transcribed.
- In the absence of glucose, intracellular cAMP levels increase, the CRP–cAMP complex is formed, and transcription of the lac operon is stimulated, allowing lactose to be used as an alternative carbon source.
- When lactose is present, lac repressor loses its capacity to bind to DNA. This allows RNA polymerase to connect to the lac operon’s promoter and initiate transcription.
- As it turns out, RNA polymerase alone does not bind the lac operon promoter particularly effectively. It might produce a few transcripts, but without the assistance of catabolite activator protein, it won’t accomplish much more (CAP).
- CAP attaches to a stretch of DNA immediately before the lac operon promoter and facilitates the attachment of RNA polymerase to the promoter, hence promoting high levels of transcription.
- CAP, like lac repressor, is encoded by a regulatory gene on the chromosome of bacteria. The CAP gene is not part of (or even close to) the lac operon.
- Constant or constitutive expression of the CAP gene. This indicates that CAP protein is constantly present in the cell to bind cAMP and “report” glucose levels to lac operon and other target genes and operons.
- CAP is not constantly active (able to bind DNA). Instead, it is controlled by cyclic AMP, a tiny molecule (cAMP). E. coli produces cAMP as a “hunger signal” when glucose levels are low.
- cAMP binds to CAP, changing its structure so that it can bind DNA and stimulate transcription. CAP cannot bind DNA and is inactive without cAMP.
- The act of transporting glucose into the cell decreases the creation of cAMP. Thus, when an abundance of glucose enters the cell, cAMP production is limited.
- If little or no glucose is available for transfer into the cell, however, cAMP synthesis is no longer blocked and cAMP levels increase. As a result: High glucose→Low cAMP; Low glucose→High cAMP
- CAP is only active at low glucose levels (cAMP levels are high). Thus, lac operon transcription is only possible at high levels in the absence of glucose. This technique ensures that bacteria activate the lac operon and begin utilising lactose only after depleting their preferred energy source (glucose).
Catabolite Repressor Protein
- Lactose is only energetically advantageous to bacteria when glucose is absent.
- Consequently, the lac operon contains two regulatory switches, one for each sugar. CRP is a protein that is utilised to assess glucose levels.
- This monitoring method is indirect and detects concentrations of the signalling chemical cAMP.
- cAMP is produced by a protein called adenylate cyclase, which is directly regulated (inhibited) by high glucose levels.
- Thus, glucose and cAMP levels are inversely connected. As glucose concentrations decline, cAMP concentrations rise, and cAMP binds to CRP. The CRP–cAMP complex has a strong affinity for target regions in the bacterial genome.
- One of the cAMP–CRP DNA target sequences is located within the promoter for lac metabolic proteins, upstream of the lac operator region.
- Note that cAMP binding increases CRP DNA binding, but sugar binding lowers DNA binding for LacI.
- DNA binding by the cAMP–CRP complex stimulates RNA polymerase transcription of lacZYA mRNA, whereas LacI binding inhibits this process.
- As a result, lactose metabolism proteins are produced at high levels only when ambient glucose levels are low and lactose levels are high.
CRP Structural Characteristics
- CRP, like LacI, is multimeric and composed of several domains.
- In this instance, the CRP is dimeric, with each monomer containing two domains.
- The N-terminal domain supports dimer formation and binds at least one cAMP molecule (two per dimer).
- In the absence of cAMP, the two monomeric DNA-binding domains are not adequately aligned for sequence-specific DNA recognition.
- In the presence of cAMP, the structure reorganises to place the two DNA-binding helix–turn–helix configurations in the correct orientation for efficient DNA binding.
- CRP–cAMP interaction also induces a DNA DNA-binding site bend, similar to LacI.
- Each monomer only attaches to one-half of the DNA-binding site, which is symmetrical along the sequence’s centre.
- Indeed, the symmetry observed in lac operon regulatory sites is a frequent characteristic of DNA regulatory binding sites, correlating with the multimeric nature of these proteins.
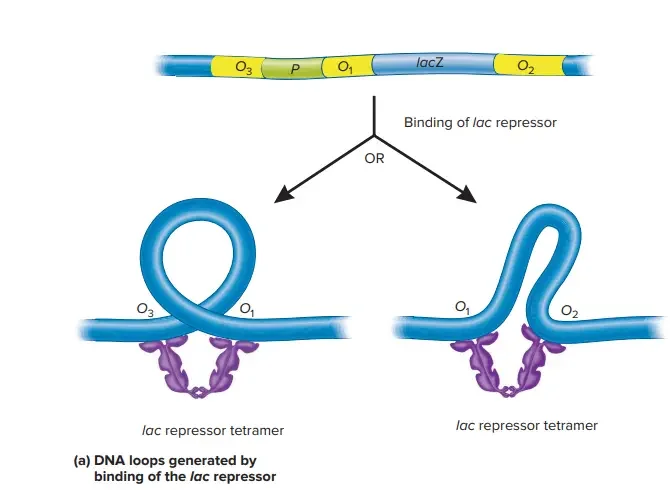
Multiple Sites/Multiple Targets – DNA Looping
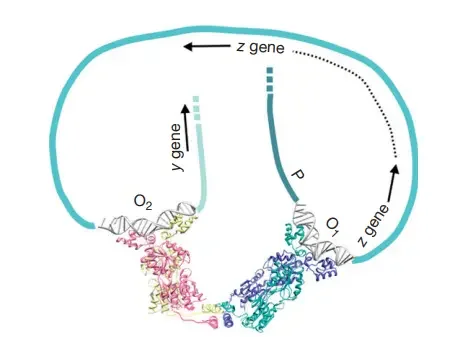
- In addition to the primary operator, LacO, the lac operon sequence contains two ‘pseudo-operator’ sequences that contribute to repression.
- The DNA sequences of the pseudo-operators are quite close, but not identical, to those of LacO, and they are weakly bound by LacI.
- The presence of two DNA-binding sites in the LacI protein tetramer revealed a way by which pseudo-operators could augment repression – a single LacI protein tetramer could bind two distinct operators and produce a DNA structure with a loop.
- Multiple laboratories have produced experimental evidence for DNA looping. Recent research suggests that the angle between two dimers must widen for loop formation to take place.
- These highly stable looping structures account for the substantial lacZYA expression suppression reported in bacterial cells.
- Indeed, DNA having numerous operator sequences and possessing the E. coli-typical supercoiling density has a complicated half-life greater than 2 days.
- However, even these looped complexes respond quickly (in less than 30 s) to the presence of inducer sugars, allowing for rapid adaptation to a temporary external lactose source.
Common Errors in Understanding the lac Operon
A number of inaccuracies are regularly observed in presentations of the lac operon
1st Error
- The most prevalent and widely disseminated misconception is that lactose is the inducer that relieves LacI suppression.
- According to this article, the molecule that functions as an inducer in vivo is allolactose, a lactose derivative produced by b-galactosidase as a byproduct of lactose hydrolysis to glucose and galactose.
- This ensures that the inducer is not created unless the metabolic capability to consume lactose is present in the cell.
2nd Error
- The second prevalent misconception is that lacZYA mRNA is never generated without lactose (and hence the inducer allolactose).
- Even in the absence of an inducer, typical dissociation events of LacI from LacO permit a little amount of read-through by RNA polymerase, resulting in extremely low levels of lacZYA mRNA production.
- This transcription ensures tiny amounts of lactose permease and b-galactosidase proteins are accessible to transport lactose and produce allolactose for induction.
- This leaky transcription can be a concern when the LacI/LacO system is employed to control the production of recombinant proteins: if the recombinant protein is toxic to E. coli, even minute amounts can be lethal.
- Thus, LacI/LacO regulation has been linked with other control systems (such as bacteriophage T7 polymerase) to impose a high level of repression until bacterial colonies reach large densities, and then to promote protein production.
3rd Error
- The third misconception is the notion that induced LacI cannot bind to the LacO operator (when bound to allolactose or gratuitous inducers).
- In reality, LacI inducer can bind to LacO, albeit with significantly lower strength.
- With this reduction in binding affinity, the LacI-inducer complex is unable to distinguish LacO from the rest of the DNA in the bacterial genome. The repressor protein is outcompeted by this surplus of nonspecific DNA, allowing RNA polymerase to transcribe lacZYA. This is because the genome is significantly larger than LacO, which represents a very, very small portion of the E. coli DNA sequence.
Conditions required to turn on lac operon
The lac operon will be highly expressed if two conditions are met:
- Glucose must be absent: When glucose is absent, cAMP binds to CAP, allowing CAP to bind DNA. Bound CAP aids in the attachment of RNA polymerase to the lac operon promoter.
- Lactose must be available: If lactose is accessible, the operator will release the lac repressor (by binding of allolactose). This permits RNA polymerase to progress along the DNA and transcribe the operon.
Together, the engagement of the activator and the release of the repressor allow RNA polymerase to bind tightly to the promoter and clear the way for transcription. They result in robust transcription of the lac operon and development of lactose-utilizing enzymes.
Lac operon Mechanism – In different Condition
Now that we’ve seen all the moving pieces of the lac operon, let’s put what we’ve learned to use by seeing how the operon responds to various situations (presence or absence of glucose and lactose).
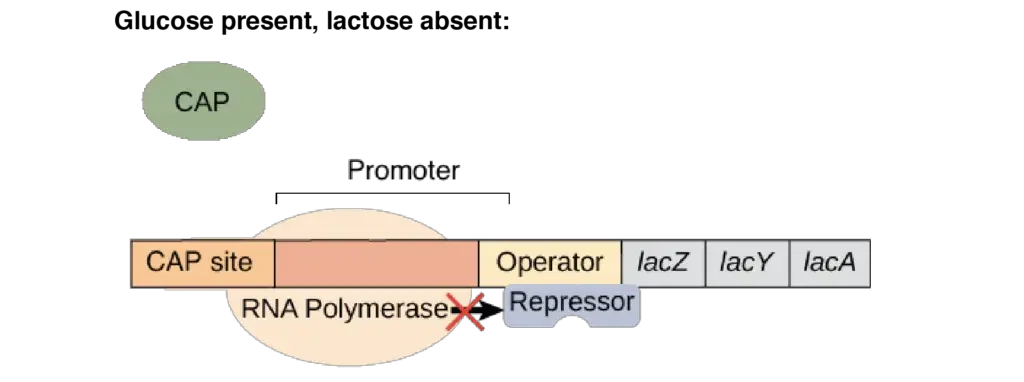
- Glucose present, lactose absent: There is no transcription of the lac operon. Because the lac repressor remains attached to the operator and blocks RNA polymerase transcription, this is the case. Also, because glucose levels are high and cAMP levels are low, CAP is inactive and cannot bind DNA.
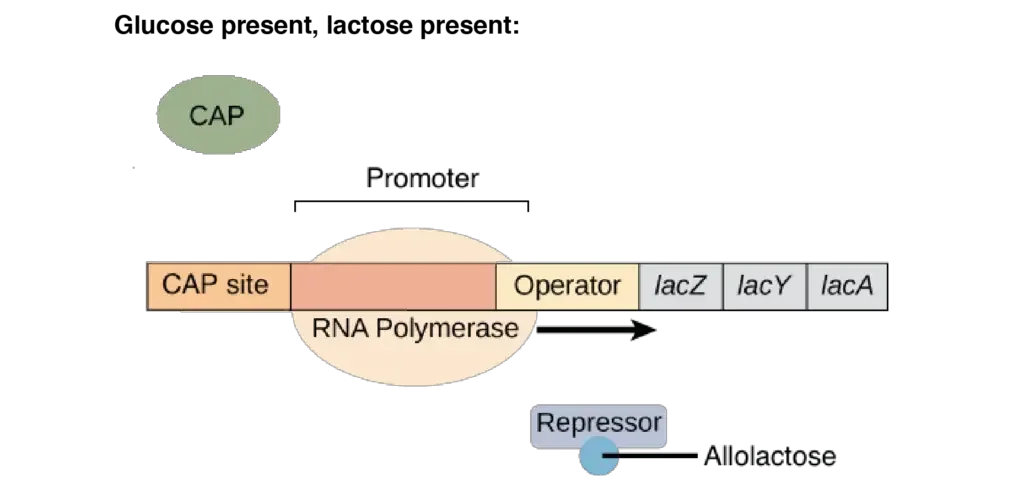
- Glucose present, lactose present: There is low-level transcription of the lac operon. Lac repressor is released from the operator due to the presence of the inducer (allolactose). cAMP levels are however low due to the presence of glucose. Thus, CAP remains inactive and is unable to bind to DNA, limiting transcription to a low, leaky level.
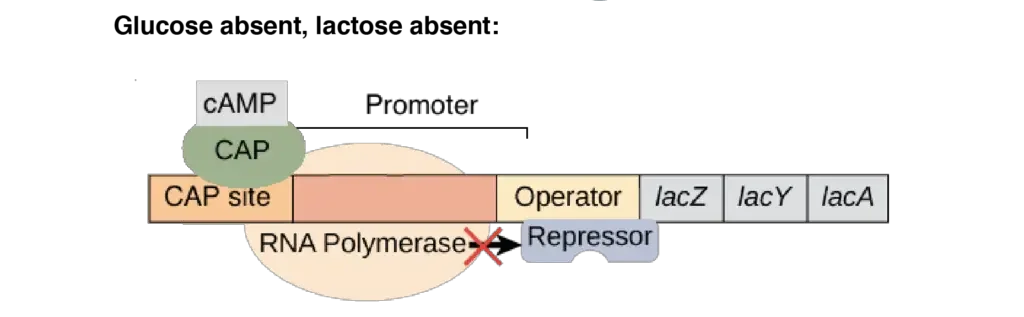
- Glucose absent, lactose absent: There is no transcription of the lac operon. Since glucose levels are low and cAMP levels are high, CAP is active and will bind to DNA. Due to the absence of allolactose, the lac repressor will also bind to the operator, functioning as a blockage to RNA polymerase and blocking transcription.
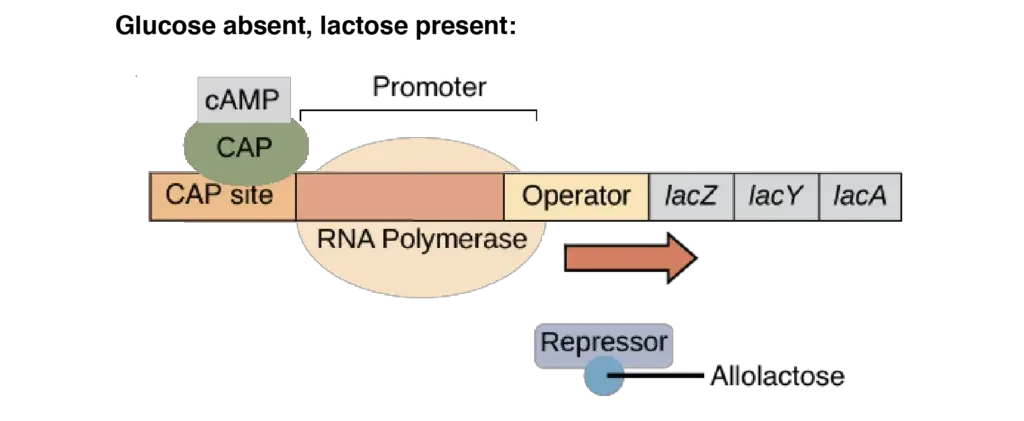
- Glucose absent, lactose present: There is robust transcription of the lac operon. Lac repressor is released from the operator due to the presence of the inducer (allolactose). Due to the absence of glucose, cAMP levels are up, therefore CAP is active and attached to DNA. CAP facilitates the binding of RNA polymerase to the promoter, allowing for high amounts of transcription.
Summary of lac operon responses
| Glucose | CAP binds | Lactose | Repressor binds | Transcription |
|---|---|---|---|---|
| + | – | – | + | No |
| + | – | + | – | Some |
| – | + | – | + | No |
| – | + | + | – | Yes |
Lactose analogs
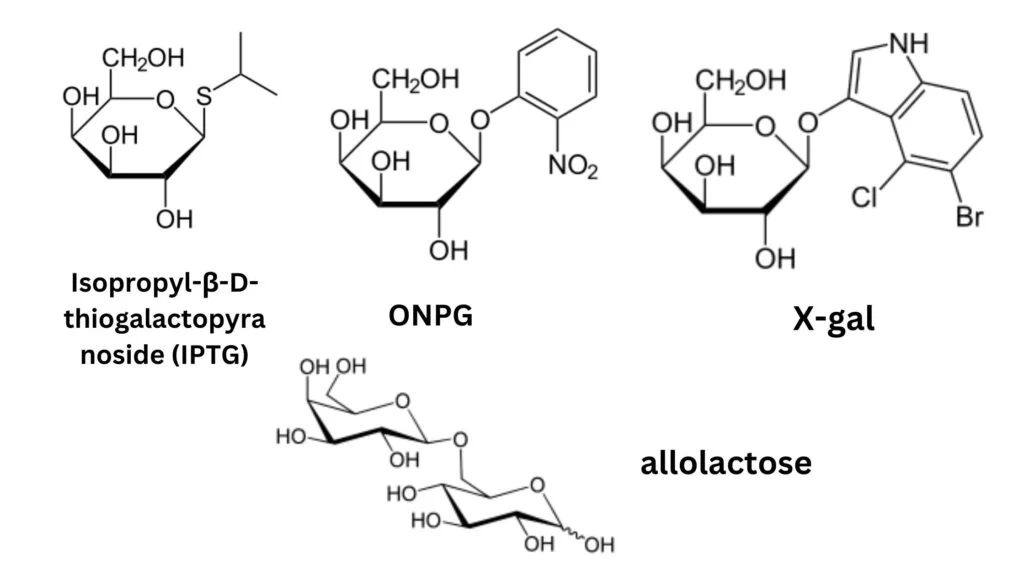
Lactose analogs, structurally modified derivatives of lactose, have garnered significant attention due to their utility in studying the lac operon, a genetic regulatory system in Escherichia coli (E. coli). These analogs are primarily substituted galactosides, wherein the glucose component of lactose is replaced by a distinct chemical entity.
- Isopropyl-β-D-thiogalactopyranoside (IPTG): IPTG stands as a prominent inducer in lac operon research. It binds to the lac repressor, rendering it inactive, yet remains unprocessed by β-galactosidase. A salient feature of IPTG is its stability in vivo; E. coli cannot metabolize it, ensuring a consistent concentration during experiments. Notably, while Pseudomonas fluorescens requires lactose permease for IPTG uptake, E. coli does not share this dependency.
- Phenyl-β-D-galactose (phenyl-Gal): Distinct from IPTG, phenyl-Gal serves as a substrate for β-galactosidase but fails to deactivate the repressor, and hence, is not an inducer. Wild-type cells, producing minimal β-galactosidase, cannot utilize phenyl-Gal as an energy source. However, mutants devoid of the repressor can metabolize it. Consequently, media containing only phenyl-Gal can selectively nurture repressor and operator mutants. Given the larger size of the lacI gene compared to the operator, repressor mutants are more prevalent.
- Thiomethyl galactoside (TMG): TMG, another lactose derivative, inhibits the lacI repressor. At minimal concentrations, both TMG and IPTG rely on lactose permease for cellular entry. Yet, at elevated levels, they can permeate cells autonomously. High extracellular TMG concentrations can impede growth rates.
- Colorimetric Indicators: Certain lactose analogs serve as visual indicators of β-galactosidase activity. For instance, ONPG, upon cleavage, yields the vibrant yellow compound, orthonitrophenol, and galactose, making it a preferred substrate for in vitro β-galactosidase assays. Similarly, X-gal produces a deep blue hue upon hydrolysis by β-galactosidase, turning colonies blue, facilitating visual differentiation.
- Allolactose: Allolactose, an isomer of lactose, acts as the natural inducer of the lac operon. While lactose has a galactose-β(1→4)-glucose structure, allolactose adopts a galactose-β(1→6)-glucose configuration. Intriguingly, β-galactosidase can convert lactose to allolactose. Demonstrating the pivotal role of LacZ in generating the genuine inducer, E. coli cells with a null lacZ mutation can produce LacY permease in the presence of IPTG but not lactose. This underscores the necessity of lactose conversion to allolactose, facilitated by β-galactosidase, to generate the intracellular inducer.
Lac operon Important Notes
- Lac operon comprises metabolically relevant genes.
- The genes are exclusively expressed in the presence of lactose and the absence of glucose.
- In reaction to glucose and lactose levels, the catabolite activator protein and lac repressor operon is turned on and off.
- The lac repressor inhibits the operon’s transcription. In the presence of lactose, its repressor function ceases.
- When glucose levels are low, catabolite activator protein initiates the transcription of the operon.
Control of Gene Expression in Prokaryotes
In prokaryotes, the Lac-operon system is controlled in two ways:
- Positive control
- Negative control
Positive Control of Lac-Operon
The positive control of the lac operon refers to the regulatory mechanism that activates gene expression when certain conditions are met. In the case of the lac operon, the presence of an inducer, such as lactose, triggers the positive control.
Here are the steps involved in the positive control of the lac operon:
- Expression of the Repressor Protein: The regulatory gene of the lac operon expresses the lac repressor protein. The repressor protein is continuously synthesized and present in the cell.
- Production of Repressor Proteins: The expression of the regulatory gene leads to the production of repressor proteins.
- Binding of the Inducer: The repressor protein has binding sites for both the operator and the inducer (lactose). When lactose is present in the cellular environment, it acts as an inducer and binds to the repressor protein.
- Formation of the R+I Complex: The binding of the inducer (lactose) with the repressor protein forms a complex called the R+I complex. This complex alters the conformation of the repressor protein.
- Prevention of Repressor Binding: The R+I complex no longer binds to the operator region. As a result, it no longer blocks the binding of RNA polymerase to the promoter region of the lac operon.
- Transcription and mRNA Production: With the repressor protein no longer blocking the operator, RNA polymerase can bind to the promoter region and initiate transcription. This leads to the production of mRNA from the lac operon genes.
By the presence of an inducer, such as lactose, the positive control of the lac operon allows for the switch-on of gene expression. The inducer prevents the repressor protein from binding to the operator, enabling RNA polymerase to transcribe the genes of the lac operon and produce the necessary enzymes for lactose metabolism.
Negative Control of Lac-Operon
The negative control of the lac operon refers to the regulatory mechanism that inhibits gene expression in the absence of an inducer, such as lactose. It involves the action of the lac repressor protein. Here are the steps involved in the negative control of the lac operon:
- Expression of the Repressor Protein: The regulatory gene of the lac operon expresses the lac repressor protein. The repressor protein is continuously synthesized and present in the cell.
- Production of Repressor Proteins: The expression of the regulatory gene leads to the production of repressor proteins.
- Binding of Repressor to Operator: In the absence of an inducer or lactose, the repressor protein binds directly to the operator region of the lac operon. This binding physically obstructs the movement of RNA polymerase and prevents its attachment to the promoter region.
- Blockage of Transcription: The binding of the repressor protein to the operator effectively blocks the transcription of the lac operon genes. RNA polymerase is unable to proceed with transcribing the mRNA.
- Switching Off the Lac Operon: In the absence of an inducer, the lac operon remains switched off. The absence of lactose as an inducer allows the repressor protein to remain bound to the operator, inhibiting gene expression.
Lac Operon Regulation by cyclic AMP
- The explanation for diauxie required the identification of additional mutations affecting the lac genes, in addition to those accounted for by the traditional hypothesis.
- Two more genes, cya and crp, were subsequently found that mapped far from lac and that, when mutated, result in a reduction in expression in the presence of IPTG and in strains of the bacteria without the repressor or operator.
- The discovery of cAMP in E. coli led to the revelation that mutants deficient in the cya gene, but not the crp gene, might have their activity restored by adding cAMP to the media.
- cAMP is produced by adenylate cyclase, which is encoded by the cya gene. In a cya mutant, the absence of cAMP reduces lacZYA gene expression by more than tenfold compared to normal.
- Adding cAMP corrects the reduced Lac expression observed in cya mutants. Catabolite activator protein (CAP) or cAMP receptor protein is encoded by the second gene, crp (CRP).
- However, lactose metabolism enzymes are produced in small amounts in the presence of both glucose and lactose (sometimes referred to as leaky expression) because the LacI repressor rapidly associates/dissociates from the DNA as opposed to tightly binding to it, allowing RNAP to bind and transcribe mRNAs of lacZYA.
- Leaky expression is required so that some lactose can be metabolised after the glucose source has been depleted, but before lac expression is fully triggered.
- The time required to manufacture sufficient numbers of lactose-metabolizing enzymes is reflected in the interval between growth phases.
- First, the CAP regulatory protein must assemble on the lac promoter, hence increasing lac mRNA output.
- Significantly more LacZ (-galactosidase, for lactose metabolism) and LacY (-galactosidase, for lactose metabolism) copies are produced when there are more lac mRNA copies (lactose permease to transport lactose into the cell).
- After a delay necessitated by the elevation of lactose-metabolizing enzymes, the bacteria undergo a new phase of fast cell division.
- Catabolite suppression presents two mysteries: first, how cAMP levels are connected to the presence of glucose, and second, why the cells bother.
- After lactose is cleaved, glucose and galactose are produced (easily converted to glucose). In terms of metabolism, lactose is an equivalent carbon and energy source to glucose.
- The quantity of cAMP is not connected to intracellular glucose concentration, but rather to the pace of glucose transport, which affects the activity of adenylate cyclase. Additionally, glucose transport inhibits lactose permease directly. As to why E. coli behaves in this manner, only speculation can be made.
- All intestinal bacteria digest glucose, indicating that they are frequently exposed to it. It may be advantageous for cells to regulate the lac operon in this manner due to a little differential in the efficiency of transport or metabolism between glucose and lactose.
In summary
- When lactose is lacking, relatively little Lac enzyme synthesis occurs (the operator has Lac repressor bound to it).
- When lactose and a chosen carbon source (such as glucose) are present, a little amount of enzyme is generated (Lac repressor is not bound to the operator).
- In the absence of glucose, CAP-cAMP binds to a particular DNA location upstream of the promoter and interacts directly with RNAP to facilitate the binding of RNAP to the promoter.
What Do You Mean by Inducers and the Induction of Lac operon?
The lac operon, a genetic regulatory system found in the bacterium Escherichia coli (E. coli), orchestrates the expression of genes essential for lactose metabolism. This system is a paradigmatic example of gene regulation, responding dynamically to environmental cues, specifically the presence or absence of lactose and glucose.
Inducers and Their Role in the Lac Operon Activation
In the typical state, E. coli produces minimal quantities of the three proteins associated with lactose metabolism. However, when lactose permeates the cellular environment, a significant upregulation of these enzymes occurs. This phenomenon, termed “induction,” is facilitated by the conversion of lactose into allolactose by the sparse ß-galactosidase molecules present. Allolactose, thus, acts as an inducer, initiating the transcription of the genes within the lac operon. Another notable inducer is isopropylthiogalactoside (IPTG), which, unlike allolactose, remains unmetabolized by E. coli, rendering it a valuable tool for experimental induction studies.
Regulation in the Absence of Inducers
Without inducers like allolactose or IPTG, the lacI gene undergoes transcription, leading to the synthesis of a repressor protein. This repressor binds to the operator site (Olac) of the lac operon, inhibiting the transcription of the lacZ, lacY, and lacA genes.
Regulation in the Presence of Inducers
Upon induction, the inducer molecule interacts with the repressor, inducing a conformational shift that diminishes its binding affinity for the lac operator site. Consequently, the repressor detaches, enabling the adjacent RNA polymerase to commence the transcription of lacZ, lacY, and lacA genes. This transcription results in a polycistronic mRNA, ensuring coordinated regulation of all three gene products. If the inducer is subsequently removed, the repressor swiftly reassociates with the operator, halting transcription.
The Role of CRP/CAP in Transcription Enhancement
Optimal transcription of the lac operon necessitates the catabolite activator protein (CAP), also recognized as the cAMP receptor protein (CRP). This dimeric protein requires complexation with 3’5′ cyclic AMP (cAMP) to bind DNA. The CRP–cAMP complex associates with the lac promoter proximal to the RNA polymerase binding site, enhancing its binding and thereby amplifying transcription.
Influence of Glucose on the Lac Operon
The lac operon’s activity is intricately linked to the cellular glucose levels. When glucose is abundant, E. coli prioritizes its utilization over lactose, rendering the lac operon redundant. Evolutionarily, this system has developed glucose sensitivity. The presence of glucose suppresses adenylate cyclase, responsible for cAMP synthesis. Consequently, intracellular cAMP concentrations drop, preventing CRP from binding to the lac promoter, resulting in subdued lac operon activity, even if lactose is available.
Conversely, in glucose’s absence, adenylate cyclase remains uninhibited, elevating cAMP levels. This ensures that, in environments devoid of glucose but abundant in lactose, the CRP–cAMP complex augments lac operon transcription, permitting lactose utilization as an alternate carbon source. However, without lactose, the lac repressor ensures operon inactivity. This dual control guarantees robust transcription of lacZ, lacY, and lacA genes only when glucose is scarce and lactose is abundant.
In summary, the lac operon exemplifies the intricate regulatory mechanisms bacteria employ to adapt to fluctuating environmental conditions, ensuring optimal energy utilization and survival.
Lac Operon In Summary
- lac operon is the cluster of genes that controls the development of the enzymes necessary for lactose degradation in bacterial cells.
- The lac operon is an example of an inducible operon, which is an operon in which the presence of a crucial metabolic component (lactose in this case) triggers transcription of the structural genes.
- The lac operon has three tandem structural genes: the z gene, which encodes -galactosidase; the y gene, which encodes galactoside permease, a protein that stimulates the entry of lactose into the cell; and the a gene, which encodes thiogalactoside transacetylase, an enzyme with an unknown physiological function.
- If lactose is present in the media, the disaccharide enters the cell and attaches to the lac repressor, altering the repressor’s conformation and preventing it from binding to the DNA of the operator. In this condition, structural genes are transcribed, enzymes are produced, and lactose molecules are catabolized.
- Therefore, in an inducible operon such as the lac operon, the repressor protein can only attach to the DNA in the absence of lactose, which works as the inducer.
- The disaccharide dissociates from its binding site on the repressor molecule when the content of lactose in the media drops.
- Lactose release permits the repressor to bind to the operator, which physically prevents the polymerase from reaching the structural genes, hence inhibiting transcription of the operon.
- Cyclic AMP Exerts Positive Control Repressors, such as those of the lac and trp operons, exercise their influence through negative control, as the interaction of this protein with the DNA reduces gene expression.
- Earlier research on a phenomenon known as the glucose effect revealed that the lac operon is also subject to positive control.
- If bacterial cells are provided with glucose plus a range of other substrates, such as lactose or galactose, the cells will catabolize the glucose while ignoring the other molecules.
- The presence of glucose in the medium inhibits the formation of certain catabolic enzymes, such as -galactosidase, required to breakdown these other substrates.
- In 1965, a startling discovery was made: cyclic AMP (cAMP), which was previously believed to be only engaged in eukaryotic metabolism, was found in E. coli cells.
- The concentration of cAMP in the cells was shown to be inversely proportional to the concentration of glucose in the medium; the higher the glucose concentration, the lower the cAMP concentration.
- In addition, when cAMP was given to the medium in the presence of glucose, the cells generated catabolic enzymes that are normally missing.
- Although the precise method by which glucose decreases the content of cAMP has not yet been elucidated, the mechanism by which cAMP counteracts this impact is well understood.
- As expected, a tiny chemical such as cAMP could not act alone to trigger the expression of a specific battery of genes.
- Similar to eukaryotic cells, cAMP exerts its effects on prokaryotic cells via binding to a protein, in this case the cAMP receptor protein (CRP).
- CRP cannot bind to DNA independently. In contrast, the cAMP–CRP complex identifies and binds to a particular location in the lac regulatory region.
- The presence of the attached CRP causes a change in the DNA’s structure, which allows RNA polymerase to transcribe the lac operon.
- Thus, even in the presence of lactose and an inactive repressor, transcription of the operon requires the presence of the bound cAMP–CRP complex.
- As long as glucose is available, cAMP levels stay below those required to stimulate transcription of the operon.
trp operon vs lac operon
The Lac operon and Trp operon are two distinct regulatory systems found in bacteria. Here are some of the key differences between the two operons:
- Function: The Lac operon is involved in the catabolism of lactose, a sugar, while the Trp operon is involved in the anabolic process of synthesizing tryptophan, an amino acid.
- Regulation by Inducer: The Lac operon is activated in the presence of lactose as an inducer. When lactose is present, it binds to the repressor protein, preventing it from binding to the operator and allowing gene expression. In contrast, the Trp operon is deactivated in the presence of tryptophan as an inducer. Tryptophan binds to the repressor protein, enabling it to bind to the operator and block gene expression.
- Number of Genes: The Lac operon consists of three structural genes (lac Z, lac Y, lac A) and a repressor gene (lac I). These genes are responsible for lactose metabolism. On the other hand, the Trp operon consists of five structural genes (trp E, trp D, trp C, trp B, trp A) and a repressor gene (trp R). These genes are involved in the synthesis of tryptophan.
- Regulatory Mechanism: The Lac operon does not utilize an attenuation mechanism for regulation. Instead, it relies on the binding of the repressor protein to the operator. In contrast, the Trp operon employs an attenuation mechanism. This mechanism involves the premature termination of transcription in the presence of high levels of tryptophan.
| Aspect | Lac Operon | Trp Operon |
|---|---|---|
| Function | Catabolism of lactose | Anabolism of tryptophan |
| Inducer Regulation | Activated by lactose | Deactivated by tryptophan |
| Genes | lac Z, lac Y, lac A, lac I | trp E, trp D, trp C, trp B, trp A, trp R |
| Regulatory Mechanism | Repressor protein binding | Attenuation mechanism |
FAQ
What is the Lac operon?
The Lac operon is a genetic regulatory system found in bacteria, such as E. coli, responsible for the transport and metabolism of lactose.
How does the Lac operon work?
The Lac operon consists of a promoter, operator, and structural genes. When lactose is present, it acts as an inducer and binds to the repressor protein, allowing RNA polymerase to transcribe the structural genes.
What are the structural genes of the Lac operon?
The structural genes of the Lac operon are lac Z, lac Y, and lac A. They encode proteins involved in lactose metabolism, including beta-galactosidase, lactose permease, and beta-galactoside transacetylase.
What is the role of beta-galactosidase in the Lac operon?
Beta-galactosidase is an enzyme encoded by lac Z that catalyzes the hydrolysis of lactose into glucose and galactose, making lactose usable as an energy source.
What is the function of lactose permease in the Lac operon?
Lactose permease, encoded by lac Y, is a membrane protein that facilitates the transport of lactose into the bacterial cell, allowing its metabolism.
What is the role of the lac I gene in the Lac operon?
The lac I gene encodes the lac repressor protein. In the absence of lactose, the repressor binds to the operator, preventing transcription of the structural genes.
How is the Lac operon regulated?
The Lac operon is regulated by both positive and negative control. In the absence of lactose, the lac repressor blocks gene expression, while in the presence of lactose, the inducer molecule binds to the repressor, allowing gene transcription.
What is the significance of the lac operon in gene regulation?
The lac operon was the first genetic regulatory system to be understood. Its study provided insights into how genes are controlled and paved the way for further understanding of gene regulation in both prokaryotes and eukaryotes.
How does glucose affect the regulation of the Lac operon?
The Lac operon is subject to catabolite repression, meaning that when glucose is present, the catabolite activator protein (CAP) is not activated, reducing the transcription of the Lac operon genes.
Can the Lac operon be found in other organisms besides bacteria?
The Lac operon is primarily found in bacteria, particularly in the Enterobacteriaceae family. However, similar regulatory mechanisms involving inducible and repressible genes are observed in other organisms, although with different specificities and control elements.
References
- David Hames and Nigel Hooper (2005). Biochemistry. Third ed. Taylor & Francis Group: New York.
- Parija S.C. (2012). Textbook of Microbiology & Immunology.(2 ed.). India: Elsevier India.
- Swint-Kruse, L., & Matthews, K. S. (2013). lac Operon. Encyclopedia of Biological Chemistry, 694–700. doi:10.1016/b978-0-12-378630-2.00249-8
- Ramos, J. L., García-Salamanca, A., Molina-Santiago, C., & Udaondo, Z. (2013). Operon. Brenner’s Encyclopedia of Genetics, 176–180. doi:10.1016/b978-0-12-374984-0.01096-2
- Lehninger Principles of Biochemistry: International Edition by David L. Nelson (Author), Michael Cox (Author)
- Molecular Biology by Robert F. Weaver (Author)
- Osbourn AE, Field B. Operons. Cell Mol Life Sci. 2009 Dec;66(23):3755-75. doi: 10.1007/s00018-009-0114-3. Epub 2009 Aug 7. PMID: 19662496; PMCID: PMC2776167.
- Swint-Kruse, L., & Matthews, K. S. (2013). lac Operon. Encyclopedia of Biological Chemistry, 694–700. doi:10.1016/b978-0-12-378630-2.00249-8
- https://bitesizebio.com/21135/the-lac-operon-explained/
- https://www.khanacademy.org/science/ap-biology/gene-expression-and-regulation/regulation-of-gene-expression-and-cell-specialization/a/the-lac-operon
- https://bitesizebio.com/21135/the-lac-operon-explained/
- https://www.osmosis.org/learn/Lac_operon
- https://www.news-medical.net/life-sciences/The-Lac-Operon-in-E-Coli.aspx
- https://biologydictionary.net/lac-operon/
- https://microbenotes.com/lac-operon/