Hybridization Probes Definition
Hybridization probes are a small nucleotide sequence that is used for the detection of complementary sequences (DNA or RNA) in a nucleic acid sample.
DNA Probes detects the complementary DNA with the help of a hybridization reaction. This is achieved by permitting the probes to base pair with the sample nucleic acid and they identifying the sample that show base pairing with the proper (hybridization).
Hybridization can occur only when the base sequence of the probe is present within the gene or DNA segment which is aimed to detect. Both DNA and RNA can be used as a probe.
DNA probes are labeled with radioisotopes, epitopes, biotin, or fluorophores to enable their detection. The size of an ideal DNA probe should be 100–10000 bases long.
Example of Probe: TaqMan® probes, Molecular Beacon probes, Cycling Probe Technology, etc.
Preparation of Hybridization Probes
A Hybridization Probe can be prepared by various techniques;
- Highly purified mRNA can be used as a probe.
- Single-stranded RNA probe can be prepared by cloning the cross ponding DNA sequence into a special vector.
- Single-stranded cDNA probe can be prepared by limiting the copying of mRNA by reverse transcriptase to only one stand.
Types of Hybridization Probes
The hybridization probe is two type such as;
- DNA probe
- RNA probe
1. DNA probe
- A DNA probe is a fragment of DNA that has a nucleotide sequence specific for the gene or chromosomal region of interest. DNA probes employ nucleic acid hybridization with specifically labeled sequences to rapidly detect complementary sequences in the test sample.
- DNA hybridization probes are mainly used for the detection of complementary DNA from the test sample.
- The size of a DNA hybridization probe varies between 100 to 10000 base pair.
- DNA probes can be prepared by using a chemically or amplification method or using the cDNA library.
Example of DNA Probe:
A. TaqMan probe:
- TaqMan probe first discovered by Kary Mullis.
- TaqMan is a type of hydrolysis probe which is used in the real-time PCR assay.
- 5’ end of this probe contains a fluorophore and in 3’ end, it contains a quencher molecule which is proximity of the fluorophore.
- TaqMan probes are used for DNA or gene quantification and gene expression studies.
Mechanism of Taqman Probe:
- Once the Taqman probe binds with its complementary sequence, then, the DNA polymerase enzyme will start to synthesize a new strand but in the same, it will remove the probe by the 3’ to 5’ exonuclease activity.
- After that the probe unquenched, and the fluorophores detached from the quencher molecule and emits the fluorescent.
- Then the TaqMan probe is hydrolyzed and it started to emit fluorescent which is measured by the detector.
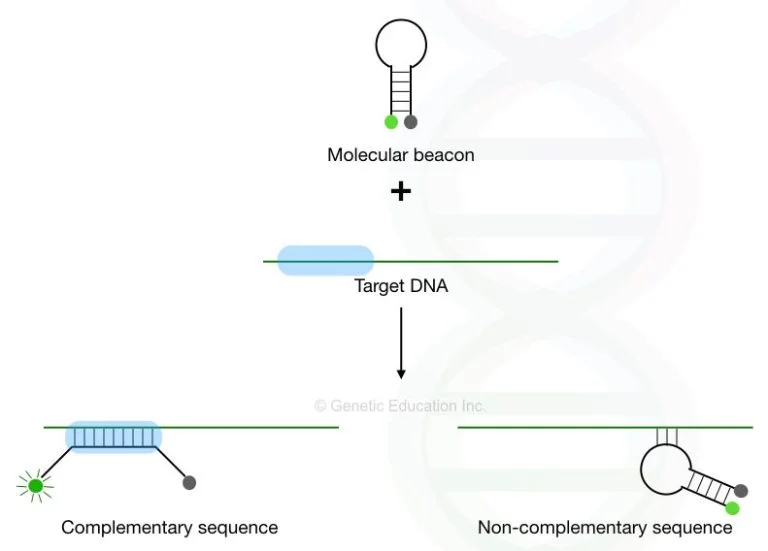
B. Molecular beacon:
- Molecular beacon is a type of hairpin probe, which is used in the quantification assay.
- Molecular beacons contain two complementary ends which are attached to each other with the help of fluorophore at one end and a quencher molecule at another end.
- The quencher dye is in close to the fluorescent molecule. That’s why the fluorophore can not emit fluorescence.
2. RNA probe
- RNA probes are stretches of single-stranded RNA used to detect the presence of complementary nucleic acid sequences (target sequences) by hybridization.
- RNA probes are usually labeled, for example with radioisotopes, epitopes, biotin or fluorophores to enable their detection.
- These probes are synthesized by in vitro transcription and can be substituted for DNA probes in nearly all applications.
- RNA probes also known as riboprobes or cRNA probes.
- RNA probes are used for the detection of mRNA in the test sample.
- Once the RNA probes are constructed In the northern blot hybridization, then it will be employed for hybridization on the nitrocellulose paper.
- Then the hybridization signal on target single-stranded nucleic acid immobilized nitrocellulose paper can be detected by using autoradiography.
Labeling of Hybridization Probes
Probe can be labeled by this two methods;
1. Radioactive labeling:
There are present various technique for labeling of nucleic acids such as;
- Primer extension.
- End labeling.
- Direct leveling.
2. Non-Radioactive labeling:
There are several strategies for non-radioactive labelling for nucleic Acid such as;
- Labeling with biotin.
- Labeling with the deoxygenin.
- Labeling with fluorescent molecule.
Probe Hybridization Steps
- At first, the taste DNA sequence is digested using restriction endonuclease.
- Then the digested fragments are allowed to immobilized on the nitrocellulose membrane.
- After that selected complementary probe is labeled and then denatured.
- The denature probes are added to nitrocellulose paper, and allowed them for binding with the complementary sequence, if there any complementary DNA present then these probes will bind with them.
- After that, the unbonded probes are removed from the nitrocellulose paper by using a wash buffer.
- Then the presence of hybridization is confirmed by using autoradiography method.
Uses or Application of Hybridization Probes
- Used in medical industries, food industries, microbial identification, and environmental science for the detection of various nucleic acid present in any biological sample.
- Identification of Recombinant clone carrying the desired DNA insert through the techniques of DNA hybridization.
- Confirmation of the integration of DNA insert into the host genome.
- Development of “Restriction fragment length polymorphism (RFLP)”.
- In situ hybridization for determining the location of a specific sequence in a specific chromosomes.
- Preparation of genome map for eukaryotes.
- Diagnosis of disease-causing by parasite pathogens virus.
Facts about Hybridization Probes
- X-ray pictures or UV light is used during the autoradiography of hybridized probes.
- The appearance chances of hybridization depends on the stringent conditions of hybridization.
- High stringency, such as high hybridization temperature and low salt in hybridization buffers, permits only hybridization between nucleic acid sequences that are highly similar.
- Whereas low stringency, such as lower temperature and high salt, allows hybridization when the sequences are less similar.
- Mainly two types of markers are used during the labeling of DNA probes such as 32P (radioactive isotope of phosphorus), and digoxigenin (It is a non-radioactive, antibody-based marker)
- The DNA probes are hybridized to a single-stranded nucleic acid (DNA or RNA) whose base sequence will allow probe-target base pairing due to the presence of complementarity between the probe and target.