Gene mapping, also known as genome mapping or genome mapping, is an important technique used by scientists to establish the location of genes on a chromosome. It entails determining the location of a gene, quantifying the distance between genes, and establishing landmarks within the genome. Just like landmarks help us make our way around a city, these genetic landmarks help researchers understand how genes are organised and how they interact with one another.
Alfred H. Sturtevant conducted pioneering research on the model organism Drosophila melanogaster in 1911, which gave rise to the concept of gene mapping. He established that genes are ordered linearly on chromosomes and determined that the distance between two genes remains constant if they are on the same chromosome and that the length of the chromosome is consistent between individuals of the species. Sturtevant also discovered that the frequency of crossing over during reproduction may be used to locate genes on a chromosome.
Sturtevant concluded from his observations that genes adjacent to each other on a chromosome are more likely to be inherited jointly, but genes farther apart are less likely to be inherited together. The genes on the same chromosome are said to be “linked,” and the distance between them is called the linkage distance.
Sturtevant’s linkage map served as the foundation for subsequent gene mapping studies, including the creation of the first human genome map. The first genetic map of the human genome, known as RFLPs (restriction fragment length polymorphisms), was published in 1987, utilising 400 distinct restriction enzyme markers.
Scientists use two basic strategies in gene mapping: genetic mapping and physical mapping. Genetic mapping is finding genes within a genome, particularly protein-coding genes of interest. Physical mapping, on the other hand, entails determining the distances between different gene locations.
Modern gene mapping initiatives frequently begin with genome sequencing, followed by computerised analysis of the resultant sequence to locate targeted genes. The ultimate goal of gene mapping is to precisely place molecular markers, such as genes, on their correct positions within the genome. Researchers can get vital insights into genetic features, inheritance patterns, and the activities of specific genes by knowing the organisation and distances between genes.
In summary, gene mapping is a critical method that scientists use to pinpoint the location of genes on a chromosome and the distances between them. It has played a critical role in understanding the organisation of genes and their interactions, allowing researchers to better comprehend genetic characteristics and inheritance patterns. Gene mapping continues to add to our understanding of the complicated world of genetics, thanks to advances in genome sequencing and digital analysis technologies.
Gene Mapping Definition
Gene mapping, also known as genetic mapping or genome mapping, is the technique of finding the relative positions and linear order of genes on a chromosome. Gene mapping offers critical information about the order, position, and distance between genes inside a genome.
Various techniques, including molecular and cytogenetic procedures, can be used to map genes. Techniques like as DNA sequencing, polymerase chain reaction (PCR), and DNA hybridization are used in molecular gene mapping to discover and locate specific DNA sequences linked with genes. These techniques enable researchers to generate genetic maps that show the physical locations of genes on chromosomes.
Cytogenetic gene mapping is the microscopic examination of chromosomes. To investigate the structure and organisation of chromosomes, techniques like as karyotyping, fluorescent in situ hybridization (FISH), and comparative genomic hybridization (CGH) are used. Researchers can establish the relative placements of genes by analysing the pattern of staining, banding, or hybridization signals on chromosomes.
Understanding the organisation of genes inside a genome, as well as investigating genetic features, inheritance patterns, and illnesses, requires gene mapping. It lays the groundwork for future study in areas such as molecular genetics, genomics, and genetic engineering. Gene mapping aids in the diagnosis, prevention, and therapy of genetic illnesses by locating the location of disease-causing genes.
Types of Gene Mapping
Genetic mapping and physical mapping are two primary types of gene mapping techniques used by scientists. Here’s a brief explanation of each:
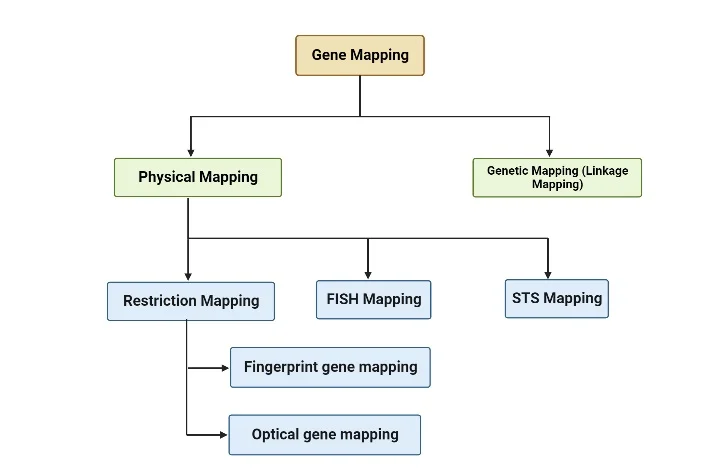
- Genetic Mapping (Linkage Mapping): Genetic mapping determines the relative positions of genes on a chromosome based on their patterns of inheritance and recombination. It relies on analyzing how genes are transmitted together or separately from one generation to the next. By studying the frequency of recombination events between genes, researchers can create a genetic map that orders genes along a chromosome. Genetic maps are measured in genetic distance units, such as centimorgans (cM). Genetic mapping is particularly useful for studying traits and diseases that exhibit genetic linkage.
- Physical Mapping: Physical mapping determines the actual physical positions of genes on a chromosome. It involves techniques that directly examine the DNA or chromosome structure. Physical mapping techniques include DNA sequencing, restriction mapping, and hybridization techniques. These methods provide information about the distances between genes and their precise locations on the chromosome. Physical maps are typically represented as linear DNA sequences, where genes are ordered and spaced according to their physical positions.
Both genetic mapping and physical mapping are important for understanding the organization and structure of genes within a genome. They complement each other, with genetic mapping providing information on the order of genes based on recombination events, and physical mapping providing the precise locations and distances between genes.
Physical mapping
Physical mapping is a gene mapping technique that determines the exact location of a gene, disease gene, or DNA sequence of interest. It provides precise information about the physical distances between genetic markers and the actual DNA base pair distances.
There are several methods used in physical mapping, and three commonly employed techniques are:
- Restriction Mapping: This method utilizes restriction enzymes, which are enzymes that cut DNA at specific recognition sequences. The DNA is digested with these enzymes, and the resulting fragments are separated using gel electrophoresis. The sizes of the fragments provide information about the distance between restriction enzyme sites on the DNA. By analyzing the pattern of DNA migration, known as the genetic fingerprint, researchers can identify specific DNA segments. This information helps in assembling contigs, which are overlapping stretches of DNA, and determining the DNA sequence of the studied organism.
- FISH Mapping (Fluorescence In Situ Hybridization): FISH mapping involves labeling specific DNA sequences with fluorescent probes and visualizing their locations on chromosomes. The probes bind to complementary sequences, allowing researchers to detect the presence and location of specific genes or DNA regions. FISH mapping provides information about the spatial arrangement of genes and helps in identifying chromosomal abnormalities and gene rearrangements.
- STS Mapping (Sequence Tagged Site Mapping): STS mapping involves identifying unique DNA sequences, known as sequence tagged sites (STS), that can serve as markers for specific locations in the genome. These STS markers are typically short DNA sequences that can be easily amplified using PCR (polymerase chain reaction). By determining the presence or absence of specific STS markers in a genome, researchers can establish their positions and create a physical map.
Physical mapping techniques, such as radiation hybrid mapping, sequence mapping, and cytogenetic mapping, are employed to produce comprehensive physical maps. These maps provide precise nucleotide numbers and physical distances between genetic markers. Physical mapping plays a crucial role in genome sequencing projects and helps identify correlations between specific DNA sequences and phenotypic features.
By fragmenting the genome into smaller pieces, characterizing and assembling these fragments, physical maps are constructed. The overlapping paths or “tiling paths” of these fragments allow researchers to infer physical distances between genomic features. This approach provides a higher level of accuracy compared to genetic maps, which are based on recombination frequencies.
Furthermore, physical mapping can be linked to genetic maps through processes like in situ hybridization, enabling the anchoring of physical map contigs onto a genetic map. This integration helps in sequencing the clones and identifying genetic markers associated with specific loci.
In summary, physical mapping is a valuable gene mapping technique that determines the precise locations and distances between genes and genetic markers on chromosomes. It aids in understanding genome structure, identifying disease-causing genes, and facilitating genome sequencing projects.
1. Restriction mapping
Restriction mapping is a gene mapping approach that involves creating a genomic map by utilising certain restriction enzymes. The method is based on the detection of restriction fragment length polymorphisms (RFLPs), which are differences in the lengths of DNA fragments caused by restriction enzyme action.
It is important to note, however, that the current mapping method is only applicable to polymorphic restriction sites. Only a tiny number of locations in the genome are highly polymorphic, making it difficult to create a thorough map of the complete genome using RFLP analysis alone.
To map unknown DNA fragments, restriction endonucleases, which are enzymes that cut DNA at specific recognition sites, are used. The digested DNA fragments are subsequently sorted by size using gel electrophoresis, either agarose gel electrophoresis or polyacrylamide gel electrophoresis (PAGE).
Depending on the kind of restriction digestion, the restriction mapping process yields two types of DNA fragments: sticky ends and blunt ends. Following digestion, the DNA fragments are sorted on the gel by size, allowing for visualisation and analysis.
Restriction mapping is frequently used to map genes on plasmids or shorter segments of DNA. However, mapping the complete genome by restriction digestion is a time-consuming, labor-intensive, and relatively imprecise process.
Different restriction endonucleases cleave the DNA at different recognition sites. The table below contains examples of recognition sites for several restriction enzymes. These recognition sites act as target sequences for restriction enzymes, allowing them to cleave the DNA at precise regions.
This technique produces a restriction map that indicates or locates all of the restriction sites present in the entire genome or a specific fragment of DNA. It contains useful information about the placement and distribution of restriction sites.
Restriction mapping is further classified into two methods:
- Fingerprint gene mapping: This method involves analyzing the pattern of DNA fragments produced by the action of restriction enzymes. The unique pattern, or “fingerprint,” of the DNA fragments can be used to identify and compare different individuals or species.
- Optical gene mapping: This method utilizes advanced imaging techniques to directly visualize and map individual DNA molecules. Optical gene mapping provides high-resolution information about the physical arrangement of DNA sequences.
a. Fingerprint gene mapping
- Fingerprint gene mapping is a technique for analysing the pattern of DNA fragments produced by certain restriction enzymes. Because it is not possible to digest the entire genome with a single restriction endonuclease and differentiate all resultant fragments on a gel, this approach is used.
- To address this issue, the genome is first broken down into smaller parts, or a bigger gene or DNA fragment is fragmented. These segments are subsequently introduced into bacterial plasmids, resulting in bacterial vector clones of DNA fragments.
- The DNA fragments contained within the clones are then digested using a specific restriction enzyme. The digested fragments are then sorted by size on an agarose gel using gel electrophoresis.
- A genomic map of the restriction site can be created by analysing the resulting fragment sizes and band patterns on the gel. The band patterns act as unique fingerprints, allowing various individuals or species to be compared and identified.
- The similarities and differences in band patterns between samples reveal whether or not specific restriction sites are present in the DNA sequence. Scientists can infer links and genetic variants by comparing many samples and identifying shared or different patterns.
- Because it allows for the examination of DNA fragment patterns and the identification of specific sequences or genetic markers, fingerprint gene mapping is a helpful tool in genetic research. This approach can reveal genetic diversity, genetic linkages, and the presence of genetic variations linked to certain traits or diseases.
- Overall, fingerprint gene mapping allows for the comparison of DNA fragment patterns generated by specific restriction enzymes, assisting in the creation of genetic maps and understanding genetic variances within populations.
b. Optical gene mapping
- Optical gene mapping is a method for directly visualising and mapping individual DNA molecules. It entails creating an optical map based on the fluorescence intensity emitted by gaps formed during DNA digestion.
- A DNA segment of interest is immobilised onto a glass slide in the first step of optical gene mapping. The DNA fragment is subsequently digested by a restriction enzyme, resulting in gaps between the digested pieces. These gaps are critical for the mapping procedure that follows.
- The slide containing the DNA fragment is dyed with a fluorescent dye after digestion. The dye bonds to the DNA, making it visible under a fluorescence microscope. The fluorescence intensity generated by the dyed DNA fragments is detected using the microscope.
- The sizes of the fluorescently labelled DNA pieces and the spaces between them can be measured by examining them under a microscope. This data is utilised to create a map of the DNA that represents the physical arrangement of the pieces.
- The optical map gives a high-resolution picture of the DNA molecule, allowing specific features such as structural changes, rearrangements, and repeating sequences to be identified. It can also aid in the detection of large-scale genomic changes such deletions, insertions, and inversions.
- Researchers can deduce the organisation and order of DNA sequences inside the mapped region by analysing fragment sizes and gaps. The optical mapping method provides vital information about the structural properties and genomic organisation.
- Optical gene mapping has various advantages over traditional mapping methods. It allows for the direct visualisation of DNA molecules, enabling for the discovery of structural differences that other techniques may overlook. Furthermore, optical mapping has a higher resolution and accuracy than gel electrophoresis procedures.
- In summary, optical gene mapping is a technology that directly visualises and maps individual DNA molecules using fluorescent labelling and microscopy. An optical map of the DNA can be created by analysing fragment sizes and gaps, providing vital insights into the genome’s organisation and structure. This approach is especially beneficial for investigating structural variants and large-scale genomic changes.
2. FISH
- Fluorescent in situ hybridization (FISH) is a technique that uses fluorescent probes to detect specific DNA sequences or disease genes within cells. In contrast to physical gene mapping methods that rely on sequence information, FISH directly visualises the presence and location of an interest DNA sequence.
- FISH artificially synthesises a short sequence of complementary DNA to the target DNA sequence or gene. A fluorescent dye is used to label this synthetic DNA sequence, known as a probe. The probe is precisely engineered to hybridise with the complementary sequence on the chromosome.
- If a labelled probe is applied to cells or tissue samples and allowed to hybridise, it will attach to its corresponding sequence if it exists on a chromosome. This binding produces a fluorescent signal, which can be seen with a fluorescent microscope. The presence and location of the DNA sequence or gene of interest on the chromosome are indicated by the fluorescent signal.
- Using numerous probes, FISH may identify multiple DNA sequences or genes at the same time. This type of FISH is known as multiplex FISH (mFISH). Researchers can differentiate and visualise numerous DNA sequences at the same time by employing distinct fluorescent dyes for each probe.
- The capacity of FISH to detect and visualise certain DNA sequences or genes has proved critical in finding disease-causing loci. FISH has played an important role in genetic research and diagnostics, allowing the discovery of chromosomal aberrations, gene rearrangements, and disease-related gene localisation. In clinical settings, commercially available probes specific to various disease-related loci are increasingly frequently used.
- In essence, FISH is a technique that detects and visualises specific DNA sequences or genes within cells by using fluorescently labelled DNA probes. The probes generate fluorescent signals by hybridising with their complementary sequences on chromosomes, which may be seen under a fluorescence microscope. FISH has helped to identify disease-causing loci and has become a valuable technique in genetic research and diagnosis.
3. STS mapping
- STS mapping (Sequence-Tagged Site mapping) is a gene mapping approach that employs polymerase chain reaction (PCR) and agarose gel electrophoresis. STS is a brief stretch of repeating DNA sequences that are present in numerous copies within a genome and are typically 100 to 500 base pairs in length.
- One of the most distinguishing features of STS is their high polymorphism, which implies that they vary between individuals or populations. Because STS sections are polymorphic, they can be easily recognised and used as genetic markers.
- The DNA sample of interest is first fragmented using restriction enzymes (RE) and then introduced into a bacterial plasmid to perform STS mapping. Within the bacterial cells, enough copies of the DNA fragments are produced.
- Once enough DNA copies have been recovered, the fragments containing the STS sections are gathered and PCR amplification is performed. PCR is a process that uses primers designed to target certain DNA sequences to amplify them.
- In the PCR procedure, the DNA sample is amplified using primers specific to the STS regions. This produces PCR amplicons, which are copies of the STS-containing DNA fragments.
- Using agarose gel electrophoresis, the PCR amplicons are separated and analysed. The amplicons are placed onto a gel matrix and exposed to an electric field in this phase. The fragments migrate through the gel according to their size, allowing them to be separated.
- The results of agarose gel electrophoresis can provide vital information on the genome’s organisation and the relationships between distinct DNA fragments. If two separate STS markers are found on the same DNA fragment, it indicates that they are physically adjacent to each other in the genome. Similarly, the presence of the same STS marker in two separate DNA fragments shows that those pieces overlap and represent contiguous areas of the genome.
- STS mapping reveals information about the physical layout of DNA sequences in the genome. The presence or absence of certain STS markers in distinct DNA fragments allows researchers to establish their relative positions and create a genome map.
2. Genetic mapping
The technique of genetic mapping is used to determine the relative positions of genes or DNA sequences on a chromosome or genome. It differs from physical mapping, which directly examines the DNA sequence related to a phenotype or disease. Using techniques such as pedigree analysis or cross-hybridization, the genetic mapping method shows the placements of related genes or DNA sequences associated with a phenotypic or disease.
Scientists typically collect blood samples from family members who have a specific disease trait as well as those who do not. These samples are then subjected to DNA extraction, and the DNA of both affected and unaffected family members is analysed. The results of this research serve as indicators for the trait or condition under investigation. The genetic map indicates the position on a chromosome of the genotype associated with a specific phenotype.
In the past, genetic mapping was largely employed to identify visible or identifiable traits in fruit flies such as eye colour, body colour, height, and wing form. However, genetic mapping is currently used for both visual and biochemical properties, allowing disease-causing genes to be mapped on a chromosome.
Several markers are used in genetic mapping techniques, including short tandem repeats (STR), which are microsatellites that are commonly used due to their prevalence on chromosomal telomeric regions. Other markers used in gene mapping include SSCP, VNTR, SNP, RFLP, and AFLP.
Recombination events are important in genetic mapping approaches because they assist determine the distance between genetic markers. Random Fragment Length Polymorphism (RFLP) is a technique that calculates the distance between two markers by measuring differences in homologous DNA sequences. Short tandem repeat polymorphisms (STRP) are commonly used in gene mapping studies focusing on single gene disorders. SNPs, as well as microsatellites and SNPs employed as markers in linkage analysis, are used in genome-wide association and linkage analysis genetic research.
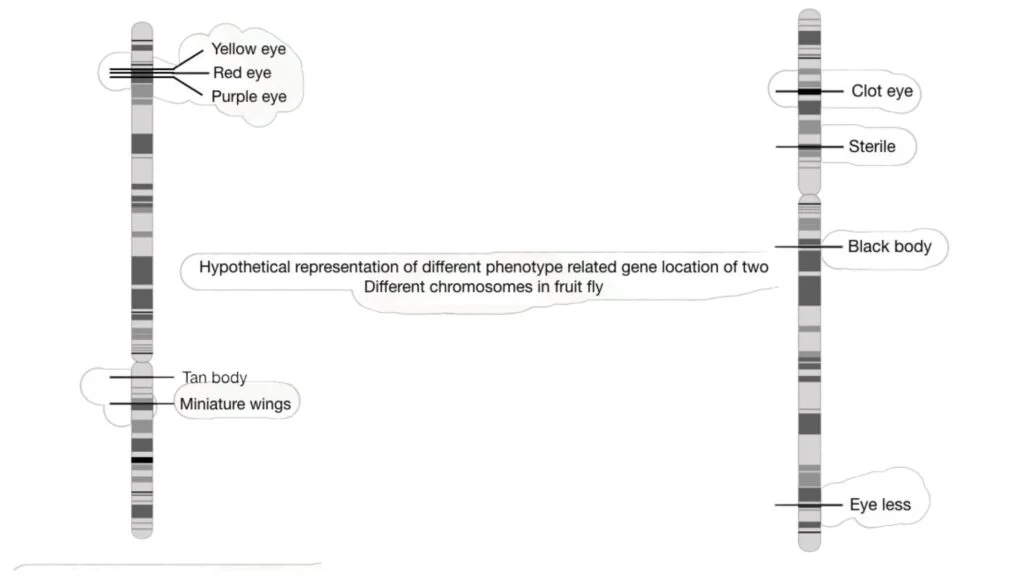
GWA studies look at the relationships between characteristics and markers like SNPs and microsatellites, treating the population as a single family. This method aids in the mapping of gene functions in common disorders.
Creating a genetic map entails creating genetic markers and a mapping population. The closer two markers are to each other on a chromosome, the more likely they will be inherited together. The order of all markers can be reconstructed by analysing their “co-segregation” patterns. The number of genetic markers and the size of the mapping population influence the quality of genetic maps. Based on their phenotypic consequences, genetic mapping can determine the relative position of genes.
Applications of gene mapping
Gene mapping, also known as genome mapping or genetic mapping, has various applications in identifying and understanding genes responsible for specific traits and diseases. Some of the important applications of gene mapping are as follows:
- Identification of Genes for Traits: Gene mapping is used to identify and locate genes responsible for specific traits in organisms. By mapping the genetic markers associated with a particular trait, researchers can determine the location of the corresponding genes on the genome. For example, in plant genetics, gene mapping can be used to identify disease-resistant genes, which can aid in developing crops with enhanced resistance to pathogens.
- Identification of Quantitative Trait Loci (QTL): Gene mapping plays a crucial role in identifying and studying quantitative trait loci (QTL). QTL are regions in the genome that are associated with complex traits such as yield, growth rate, or disease susceptibility. By mapping the genomic regions associated with these traits, researchers can gain insights into the genetic factors contributing to their variation and use this information for breeding programs or crop improvement.
- Mapping Genes in Animal Agriculture: Gene mapping is also used in animal agriculture to identify genes associated with economically important traits. For instance, mapping the genes responsible for milk production in livestock animals can help breeders select animals with higher milk yields, leading to improved productivity in the dairy industry.
- Identification of Disease-Causing Genes: In human genetics, one of the significant applications of gene mapping is the identification of disease-causing genes. By studying the genetic variations and mapping them to specific regions of the genome, researchers can pinpoint genes that contribute to the development of various diseases. This information is crucial for understanding the genetic basis of diseases and can aid in diagnosis, treatment, and prevention strategies.
- Mapping Cancer-Associated Genes: Gene mapping is valuable in the identification of cancer-associated genes, both heritable and non-heritable. By studying the genetic alterations present in cancer cells and comparing them to normal cells, researchers can identify the genes involved in the development and progression of different types of cancer. This knowledge can contribute to improved cancer diagnosis, prognosis, and the development of targeted therapies.
FAQ
What is gene mapping?
Gene mapping is a technique used to determine the relative positions of genes or DNA sequences on a chromosome or genome.
Why is gene mapping important?
Gene mapping helps in identifying and understanding the genetic basis of traits, diseases, and their inheritance patterns. It provides insights into the location and function of genes in an organism’s genome.
How is gene mapping performed?
Gene mapping involves collecting DNA samples from individuals with and without a specific trait or disease, analyzing the samples for genetic markers, and determining the linkage between markers and the trait or disease of interest.
What are genetic markers?
Genetic markers are specific DNA sequences or variations that can be used to identify and track the presence or absence of a particular gene or trait. They are used as landmarks in gene mapping.
What are the different types of genetic markers used in gene mapping?
Commonly used genetic markers in gene mapping include short tandem repeats (STRs), single nucleotide polymorphisms (SNPs), restriction fragment length polymorphisms (RFLPs), and microsatellites.
What are the applications of gene mapping?
Gene mapping is used to identify genes responsible for traits, study the genetic basis of diseases, map quantitative trait loci (QTLs), improve agricultural practices, and develop targeted therapies for genetic disorders and cancers.
Can gene mapping be used in human genetics?
Yes, gene mapping plays a crucial role in human genetics. It helps in identifying disease-causing genes, studying the inheritance patterns of genetic disorders, and understanding the genetic factors contributing to complex diseases.
How does gene mapping contribute to agriculture?
In agriculture, gene mapping aids in identifying genes associated with desirable traits such as disease resistance, yield, and quality. This information is used to develop improved crop varieties and livestock breeds through selective breeding.
What are the challenges in gene mapping?
Gene mapping can be challenging due to the complexity of the genome, the presence of repetitive DNA sequences, and the need for large and diverse populations to obtain reliable results. Additionally, technological advancements and computational tools are continuously evolving to enhance the accuracy and efficiency of gene mapping.
How has gene mapping advanced in recent years?
Advancements in technologies such as next-generation sequencing and high-throughput genotyping have revolutionized gene mapping. These technologies enable researchers to analyze large amounts of genetic data quickly and accurately, facilitating more comprehensive and detailed gene mapping studies.
certified