What is gene expression?
- Gene expression refers to the process by which information encoded in a gene is used to produce a functional gene product, typically a protein. Genes are segments of DNA that contain the instructions or code for making specific proteins. However, not all genes are active or expressed at all times in a cell. Gene expression is the regulated process through which specific genes are turned on or off, and their instructions are transcribed into messenger RNA (mRNA) molecules, which are then translated into proteins.
- The process of gene expression involves multiple steps. It begins with the activation of a gene, where specific regulatory sequences in the DNA are recognized and bound by proteins called transcription factors. These transcription factors recruit the necessary molecular machinery to initiate transcription, which is the synthesis of an mRNA molecule that carries a complementary copy of the gene’s DNA sequence.
- During transcription, the DNA double helix is unwound, and an enzyme called RNA polymerase synthesizes an mRNA molecule by incorporating complementary RNA nucleotides. The resulting mRNA molecule is a single-stranded copy of the gene’s DNA sequence.
- Once the mRNA molecule is synthesized, it undergoes further processing, including the removal of non-coding regions called introns and the addition of a protective cap and tail. These modifications, known as mRNA processing, prepare the mRNA molecule for export from the nucleus to the cytoplasm, where protein synthesis occurs.
- In the cytoplasm, the mRNA molecule serves as a template for translation, the process by which proteins are synthesized. Ribosomes, the cellular machinery responsible for protein synthesis, bind to the mRNA and read its sequence in groups of three nucleotides called codons. Each codon specifies a particular amino acid, and as the ribosome moves along the mRNA, it recruits the appropriate amino acids and joins them together to form a polypeptide chain. The polypeptide chain folds into a specific three-dimensional structure to become a functional protein.
- Gene expression is tightly regulated and can be influenced by various factors, including environmental cues, developmental stage, and cell type. Cells have mechanisms to control when and where genes are expressed, allowing them to adapt to changing conditions and perform specialized functions. Dysregulation of gene expression can lead to various diseases, including cancer and genetic disorders.
- In summary, gene expression is the process by which genetic information is transcribed into mRNA and translated into proteins. It involves the activation of specific genes, transcription of their DNA sequences into mRNA, processing and export of the mRNA to the cytoplasm, and translation into functional proteins. Gene expression is a fundamental process in biology that underlies the diversity and functionality of cells and organisms.
What is Gene Expression Profiling?
- Gene expression profiling is a powerful technique used in molecular biology to understand the activity and behavior of genes within a cell or tissue. It involves measuring the expression, or activity, of thousands of genes simultaneously, providing a comprehensive overview of cellular function.
- By analyzing gene expression profiles, researchers can gain insights into various biological processes and phenomena. For example, these profiles can distinguish between different cell types, reveal cellular responses to specific treatments or stimuli, and provide information about the cell’s environment and state.
- There are several technologies used to generate the data necessary for gene expression profiling. DNA microarrays are commonly used to measure the relative activity of known target genes. These arrays contain probes that bind to specific gene sequences, allowing researchers to quantify their expression levels. Another technique, RNA-Seq, provides information on both gene sequences and their expression levels, enabling a more comprehensive analysis.
- Gene expression profiling complements genome sequencing, as it reveals what genes are actively transcribed and expressed in a particular cell at a given time. While the genome sequence provides information about the potential functions of a cell, the expression profile reveals what the cell is actually doing. Gene expression is regulated in a highly dynamic manner, with different genes being turned “on” or “off” depending on various factors such as time of day, cellular division, local environment, and intercellular signals.
- By comparing the relative expression levels of genes in different experimental conditions, researchers can identify genes that are differentially expressed. Changes in the expression of specific genes can provide insights into cellular responses, homeostatic mechanisms, or pathological conditions. For example, higher levels of mRNA coding for a specific enzyme may indicate a cellular response to increased levels of a particular compound. Similarly, differences in gene expression between cancer cells and normal cells can help identify potential therapeutic targets for treating cancer.
- Gene expression profiling also has applications in drug development and toxicity assessment. By examining changes in gene expression patterns, researchers can evaluate the effects of drugs on cellular function and identify potential biomarkers of drug metabolism or toxicity.
- In summary, gene expression profiling is a powerful tool in molecular biology that allows researchers to simultaneously measure the activity of thousands of genes. It provides valuable insights into cellular function, differentiation, responses to stimuli, and disease mechanisms. With ongoing advancements in technology and data analysis methods, gene expression profiling holds promise as an important diagnostic tool in various fields of medicine and research.
Gene Expression Profiling Definition
Gene expression profiling (GEP) is a technology for identifying the genes that are active in a given sample of cells or tissue. This technique enables profiling of genes that are differentially expressed in disease states; thereby providing diagnostic and prognostic information.
Tools for Gene Expression Profiling
Gene expression profiling techniques
Gene expression profiling techniques are used to measure the activity levels of genes in a cell or tissue at a given time. These techniques provide valuable information about which genes are being transcribed into messenger RNA (mRNA) and, by extension, which genes are actively being expressed.
There are several commonly used gene expression profiling techniques, each with its own strengths and limitations. Here are some of the main techniques:
- Microarrays: Microarray technology is one of the earliest and widely used methods for gene expression profiling. It involves immobilizing thousands of DNA or RNA probes on a solid surface (such as a glass slide or a microchip). These probes are complementary to specific genes or gene regions of interest. Target mRNA molecules from the sample are labeled with fluorescent dyes and hybridized to the probes on the microarray. The fluorescence intensity is then measured, which indicates the relative abundance of the corresponding mRNA in the sample.
- RNA-Seq: RNA sequencing (RNA-Seq) is a high-throughput sequencing-based technique that provides a comprehensive and quantitative view of the transcriptome. In RNA-Seq, RNA molecules are isolated from the sample, converted to complementary DNA (cDNA), and sequenced using next-generation sequencing platforms. The resulting sequences are then aligned to a reference genome or transcriptome, and the abundance of each transcript can be quantified. RNA-Seq offers superior sensitivity, dynamic range, and the ability to detect novel transcripts compared to microarrays.
- Quantitative PCR (qPCR): Quantitative PCR is a widely used method to measure the expression levels of specific genes with high sensitivity and specificity. It relies on the amplification of target DNA using specific primers and detection of the amplified product in real-time using fluorescent dyes or probes. qPCR can provide absolute or relative quantification of gene expression and is often used for validation of results obtained from other profiling techniques due to its accuracy and reliability.
- NanoString: NanoString technology uses a digital color-coded barcode system to quantify gene expression levels. This technique utilizes target-specific probes that are hybridized to the mRNA molecules of interest. The probes carry a unique barcode, which can be counted and quantified using a specialized instrument. NanoString offers high specificity, sensitivity, and the ability to analyze a moderate number of genes (up to a few hundred) in a single assay.
- Single-Cell RNA-Seq: Single-cell RNA sequencing is a rapidly evolving technique that allows gene expression profiling at the single-cell level. It enables the characterization of gene expression patterns in individual cells, providing insights into cellular heterogeneity and cell-to-cell variability. Single-cell RNA-Seq involves isolating and processing individual cells, followed by cDNA synthesis and sequencing. This technique is highly valuable for understanding complex biological systems and uncovering rare cell populations.
These are just a few examples of gene expression profiling techniques commonly used in research. Each technique has its advantages and limitations, and the choice of method depends on the research goals, sample availability, cost considerations, and desired level of resolution. Integrating multiple techniques can provide a more comprehensive understanding of gene expression dynamics in various biological contexts.
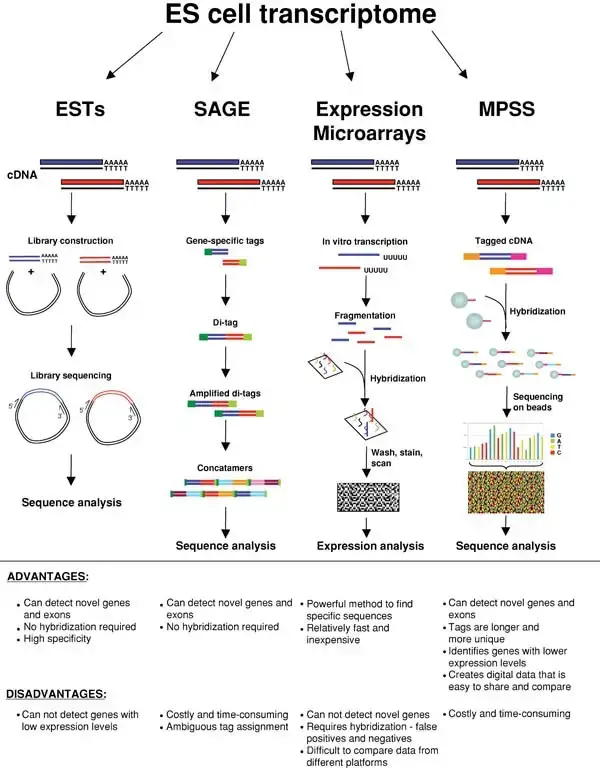
Gene expression profiling techniques used in studies of pluripotent cells
Gene expression profiling techniques have been widely employed in the study of pluripotent cells, such as embryonic stem (ES) cells, to gain insights into their unique transcriptional profiles. Here are some gene expression profiling techniques commonly used in pluripotent cell studies:
- Expressed Sequence Tag (EST) Analysis: Genome-wide expression profiling can be performed using EST analysis. In this technique, cDNA is generated from the RNA of pluripotent cells and inserted into cloning vectors to create a cDNA library. The inserts are then sequenced, and the resulting EST sequences are analyzed to identify expressed genes.
- Serial Analysis of Gene Expression (SAGE): SAGE involves the generation of short gene-specific tags, typically 10-14 base pairs in length. These tags are ligated together to form di-tags, which are then amplified by PCR, ligated again to form concatamers, and finally sequenced. SAGE allows for quantitative and comprehensive analysis of gene expression patterns in pluripotent cells.
- DNA Microarrays: Expression microarrays are widely used in gene expression profiling studies. These microarrays consist of synthesized oligonucleotides or spotted cDNAs arranged in a predetermined spatial orientation. Total RNA from pluripotent cells is reverse transcribed, fluorescently labeled, and hybridized to the microarray. The fluorescent signals generated during hybridization are detected by a scanner, and gene expression levels can be inferred from the intensity of the signals.
- Massively Parallel Signature Sequencing (MPSS): MPSS involves the cloning of a cDNA library on beads, followed by the acquisition of short 17-20 nucleotide tags from these cDNAs using an unconventional sequencing method that enables large-scale sequencing. MPSS provides a quantitative assessment of gene expression in pluripotent cells.
- Tiling Arrays: Tiling arrays are modified microarrays that represent all non-repetitive genomic DNA at various sequence resolutions. Unlike conventional microarrays, tiling arrays allow for unbiased examination of the genome, enabling the discovery of novel transcribed sequences and regulatory elements. They provide a comprehensive view of the transcriptome in pluripotent cells.
- Next-Generation Sequencing (NGS): Recently developed deep sequencing approaches using platforms such as 454 and Solexa (now Illumina) have revolutionized gene expression profiling. NGS technologies offer high-throughput sequencing, allowing for a comprehensive description of the pluripotent transcriptome. These techniques provide unprecedented sensitivity and resolution, facilitating the identification of novel transcripts, alternative splicing events, and non-coding RNAs.
By utilizing these gene expression profiling techniques, researchers can unravel the intricate transcriptional networks and regulatory mechanisms that underlie pluripotency in cells, providing valuable insights into their biological properties and potential applications.
Why use gene expression profiling?
Gene expression profiling is used for a variety of purposes in biological and medical research. Here are some key reasons why gene expression profiling is valuable:
- Understanding Cellular Function: Gene expression profiling provides a comprehensive view of the activity of thousands of genes within a cell or tissue. By measuring the expression levels of genes, researchers can gain insights into cellular functions, processes, and pathways. It helps uncover the molecular mechanisms underlying various biological phenomena.
- Disease Research: Gene expression profiling is widely used in studying diseases to identify genes and molecular signatures associated with specific conditions. By comparing gene expression profiles between healthy and diseased samples, researchers can discover genes that are differentially expressed, potentially serving as biomarkers for disease diagnosis, prognosis, and treatment response. It aids in understanding disease mechanisms, identifying therapeutic targets, and developing personalized medicine approaches.
- Drug Discovery and Development: Gene expression profiling plays a crucial role in drug discovery and development. By analyzing gene expression patterns in response to different drugs or treatments, researchers can assess their efficacy, toxicity, and mechanisms of action. It helps identify potential drug targets, predict drug responses, and optimize drug dosing and combinations.
- Biomarker Discovery: Gene expression profiling enables the identification of molecular markers or biomarkers associated with specific conditions. Biomarkers can be used for early disease detection, monitoring disease progression, predicting treatment response, and stratifying patient populations in clinical trials. Gene expression profiling aids in the development of diagnostic tests and personalized medicine approaches.
- Cell Differentiation and Development: Gene expression profiling helps to understand cell differentiation and development processes. By comparing expression profiles across different cell types or developmental stages, researchers can identify genes involved in cellular specialization and tissue formation. It provides insights into the regulatory networks and molecular events driving cellular differentiation and development.
- Environmental and Toxicological Studies: Gene expression profiling is used to assess the effects of environmental factors, toxins, and chemicals on living organisms. By examining changes in gene expression patterns, researchers can identify genes and pathways affected by environmental exposures, evaluate toxicological responses, and develop strategies for environmental monitoring and risk assessment.
Overall, gene expression profiling is a powerful tool for studying cellular processes, unraveling disease mechanisms, identifying biomarkers, and facilitating drug discovery. It provides a global perspective on gene activity and enables researchers to make informed decisions and advancements in various fields of biology and medicine.
Limitation of Gene expression profiling
Gene expression profiling, although a powerful tool in molecular biology and genomics research, has several limitations that impact its effectiveness and interpretation. Here are some key limitations of gene expression profiling:
- Differential Gene Expression: Gene expression profiling studies often focus on genes that exhibit statistically significant differences in expression levels under specific experimental conditions. However, this approach only captures a small fraction of the entire genome. Many genes are naturally turned off in different cells and tissues as a result of cellular differentiation, leading to their exclusion from expression profiles.
- Essential Genes and Survival Requirements: Numerous genes encode proteins that are vital for cellular survival and function optimally within specific concentration ranges. Consequently, the expression levels of these genes may not significantly change under experimental conditions, resulting in their exclusion from the gene expression profiles.
- Post-Transcriptional Regulation: Cells employ various mechanisms to regulate protein levels beyond altering mRNA amounts. These post-transcriptional regulatory mechanisms can lead to consistent gene expression even when protein concentrations are fluctuating. Consequently, changes in mRNA levels may not accurately reflect changes in protein expression.
- Statistical Power and Sample Size: Financial constraints often limit the number of observations for gene expression profiling experiments, resulting in a small sample size. This limited sample size reduces the statistical power of the study, making it challenging to detect subtle but important changes in gene expression.
- Biological Significance: Analyzing the biological significance of each regulated gene requires considerable effort and expertise. Due to the extensive number of genes profiled, scientists often focus their discussion on a subset of genes, leading to a potential oversight of important findings. Attaching biological significance to expression profiling results remains a complex problem, although newer techniques and databases are automating certain aspects of this process.
- Lack of Consensus: The relatively short gene lists published from expression profiling experiments limit the ability to achieve consensus among studies conducted in different laboratories. Without a comprehensive view of gene expression patterns, comparing and reconciling results from different experiments becomes challenging.
- Publicly Accessible Databases: Publishing expression profiling results in publicly accessible microarray databases allows researchers to assess expression patterns beyond what is reported in individual publications. This enables the identification of similarities between their own work and existing datasets, promoting further understanding and knowledge sharing.
In conclusion, while gene expression profiling provides valuable insights into gene regulation, it is important to consider its limitations. Researchers should be aware of the inherent biases and constraints associated with gene expression profiling techniques to ensure accurate interpretation and integration of results.
FAQ
What is gene expression profiling?
Gene expression profiling is a technique used to measure the activity levels of genes in a cell or tissue at a given time. It provides insights into which genes are being transcribed and expressed.
Why is gene expression profiling important?
Gene expression profiling helps us understand how genes are regulated and how they contribute to biological processes and disease. It allows researchers to identify genes involved in specific cellular functions, pathways, or diseases.
What are the commonly used methods for gene expression profiling?
Common methods include microarrays, RNA sequencing (RNA-Seq), quantitative PCR (qPCR), and digital PCR. These techniques provide different levels of sensitivity, resolution, and throughput.
How do microarrays work for gene expression profiling?
Microarrays consist of thousands of DNA probes that hybridize to complementary target RNA molecules. The intensity of the fluorescent signals generated during hybridization provides information about gene expression levels.
What is RNA-Seq and why is it widely used?
RNA-Seq is a high-throughput sequencing-based technique that provides a comprehensive view of the transcriptome. It offers superior sensitivity, dynamic range, and the ability to detect novel transcripts compared to microarrays.
How is gene expression profiling used in disease research?
Gene expression profiling can identify genes that are differentially expressed in disease states, providing potential diagnostic or prognostic markers. It can also help uncover underlying molecular mechanisms and identify therapeutic targets.
Can gene expression profiling be used to study single cells?
Yes, single-cell RNA sequencing (scRNA-Seq) allows gene expression profiling at the individual cell level. It provides insights into cellular heterogeneity, cell types, and developmental trajectories.
What are the challenges in gene expression profiling?
Challenges include data preprocessing and normalization, addressing technical variation, sample heterogeneity, choosing appropriate statistical analysis methods, and interpreting the biological significance of differentially expressed genes.
How can I analyze gene expression profiling data?
There are various bioinformatics tools and software packages available for gene expression data analysis. These tools assist in data preprocessing, differential expression analysis, functional enrichment analysis, and visualization of expression profiles.
What are the applications of gene expression profiling?
Gene expression profiling has broad applications, including biomarker discovery, drug target identification, understanding developmental processes, studying disease mechanisms, and personalized medicine. It is a versatile tool for advancing our understanding of gene regulation and cellular functions.
References
- Grskovic M, Ramalho-Santos M. The pluripotent transcriptome. 2008 Oct 10. In: StemBook [Internet]. Cambridge (MA): Harvard Stem Cell Institute; 2008-. Figure 1, Gene expression profiling techniques used in studies of pluripotent cells. Available from: https://www.ncbi.nlm.nih.gov/books/NBK27065/figure/thepluripotenttranscriptome.F1/ doi: 10.3824/stembook.1.24.1
- https://lab.vanderbilt.edu/miller-lab/research/gene-expression-profiling/
- https://www.science.org/doi/10.1126/science.1072241
- https://www.creative-biolabs.com/drug-discovery/therapeutics/gene-expression-profiling.htm
- https://www.htgmolecular.com/blog/2022-07-08/introduction-to-gene-expression-profiling