What is Gal operon?
The galactose operon is the unit in bacteria that is responsible for the metabolism of D-galactose, and it is organized with four structural genes (galE, galT, galK and galM). It is the process where these genes are required both for catabolic breakdown of galactose and for anabolic formation of compounds like UDP–galactose which is needed for different cellular functions.
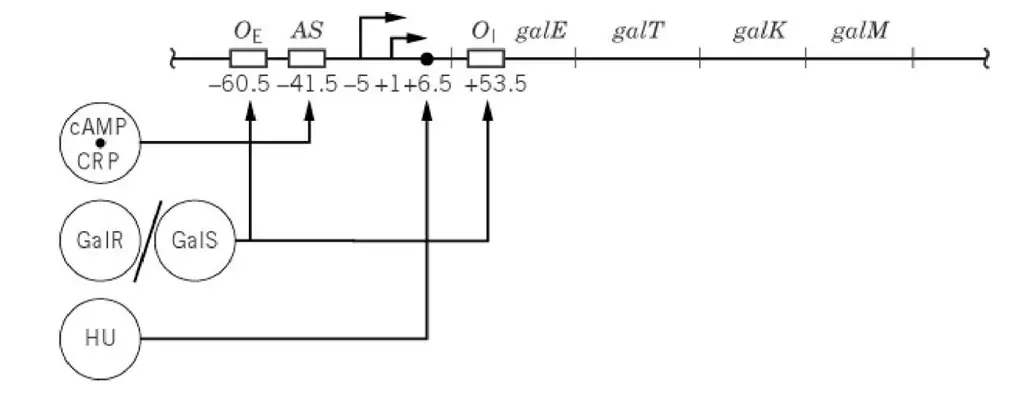
The operon is transcribed from two overlapping promoters known as P1 and P2, and these start sites are indicated as +1 and –5, and it is the arrangement that allows differential regulation in changing growth phases. The expression is controlled by regulatory proteins, and among the important is the Gal repressor (GalR) which binds with two operators, the external operator located upstream and the internal operator located after galE, and when both operators is bound a DNA loop is formed.
It is the loop that interferes with the attachment of RNA polymerase to the promoter, and this is referred to as strong repression of transcription. It is also known that this looping needs the architectural protein HU and negatively supercoiled DNA.
The operon also responds to CRP–cAMP complex which is a regulator of catabolite control. It is observed that this complex stimulates P1 promoter for higher transcription during galactose catabolism, but it inhibits P2, and this is how the cell balances catabolic and anabolic requirements. GalR shows the opposite pattern because it inhibits P1 and stimulates P2.
It is the distribution of promoter activity that changes with growth conditions, because during exponential growth P1 produces most of the transcripts, whereas during stationary phase P2 becomes the major promoter.
In this way the gal operon is seen as a system that uses two promoters and two operators to coordinate enzyme synthesis for both energy generation and biosynthetic pathways in bacteria.
Structure of Gal operon
- The gal operon is the unit that controls the utilisation of D-galactose in E. coli. It is the process where four genes is arranged in a single transcriptional cluster.
- The structural genes is placed in a sequence. These are–
- galE encoding UDP-galactose 4′-epimerase which is required for UDP-galactose formation for cell wall components.
- galT encoding galactose transferase that transfer the galactose group to an acceptor.
- galK encoding galactokinase which phosphorylate α-D-galactose to galactose-1-phosphate.
- galM encoding mutarotase that convert β-D-galactose into α-D-galactose in the first step.
- Two promoters is present. These are the overlapping promoters P1(+1) and P2(–5). It is the sites where transcription is started.
- Two operator sites is found – the external operator OE near the promoter region and the internal operator OI just after galE gene. These operators bind GalR repressor and the DNA is looped during repression.
- A histone-like protein binding site (HU site) is located between OE and OI. It is required for forming the stable DNA loop.
- An activating site (AS) is present between the external operator and the P2 promoter. This is the binding region for the cAMP-CRP complex which activate P1 and reduce the activity of P2.
- The overall structure is arranged in a way so that repression and activation occur together depending on galactose presence and the carbon source condition.
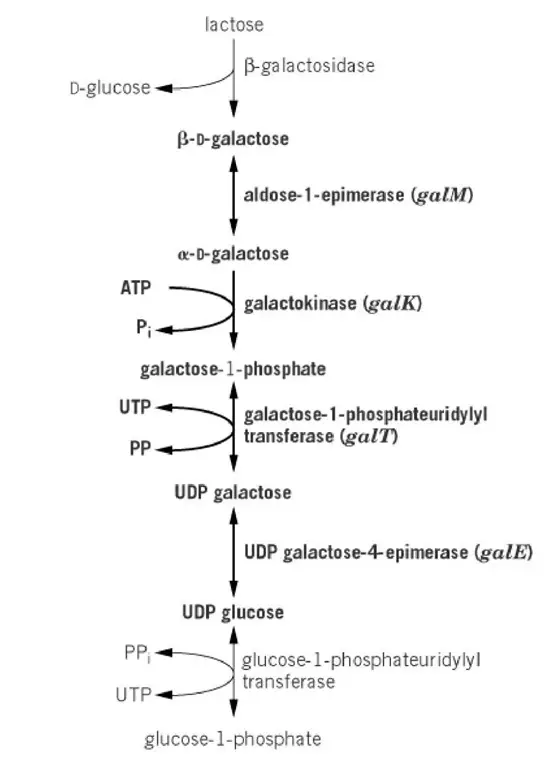
The Leloir pathway of D-galactose metabolism
- It is the main pathway for D-galactose utilisation inside the cell. The galactose is obtained either by import through permeases or by hydrolysis of lactose in the cytoplasm.
- The pathway begins with the formation of β-D-galactose from lactose. This is produced when lactose is hydrolysed, and this sugar cannot be directly phosphorylated.
- In this step the β-D-galactose is changed into α-D-galactose. This is referred to as mutarotation. It is catalysed mainly by aldose-1-epimerase (galM enzyme). The spontaneous conversion also occur but at a slow rate.
- The α-D-galactose is now acted upon by galactokinase (galK). It is the process where α-D-galactose is phosphorylated and forms galactose-1-phosphate.
- The next reaction is controlled by galactose transferase (galT). Here the galactose-1-phosphate is transferred to UDP-glucose and forms UDP-galactose and glucose-1-phosphate.
- UDP-galactose 4′-epimerase (galE) converts UDP-galactose to UDP-glucose. These two nucleotide sugars is needed for producing complex carbohydrates.
- These enzymes is connecting catabolism with biosynthesis. It is the pathway which supply UDP-glucose and UDP-galactose for cell wall components and also break down the galactose for energy.
- The whole pathway therefore join lactose utilisation and galactose utilisation into a single metabolic route by the activity of aldose-1-epimerase.
Regulation of Transcription
- The gal operon is transcribed from two promoters P1 and P2. These promoters is separated by only 5 bp which make them act in a coordinated manner.
- The main regulator is the Gal repressor GalR. It is the protein that bind to the operator regions and keep the transcription at a low level.
- The histone-like protein HU works as a corepressor. When HU binds with GalR, the DNA is looped and repression is strengthened.
- In the presence of D-galactose or non-metabolizable analogs like D-fucose, the repression is relieved. It is the process where the inducer binds GalR and change its conformation.
- This allosteric effect decrease the binding of GalR to DNA and the transcription becomes increased up to 15-fold. It is still not clear whether α- or β-anomer or both act as the actual inducer.
- Both P1 and P2 promoters is controlled by GalR, by GalS (a Gal isorepressor), and by the cAMP–CRP complex. These regulators influence the promoters in opposite directions.
- The activity of the promoters is also affected by cis-acting DNA regions. These sequences act independently without needing regulatory proteins.
- The overall regulation therefore depend on repressor binding, inducer binding, promoter arrangement and the cAMP–CRP complex that respond to carbon source conditions.
Regulation without Regulatory Proteins
Control of P1 by Adenine Tracks
- In the absence of regulatory proteins the P1 and P2 promoters show almost equal intrinsic strength. Both are modestly active during basal expression.
- It is found that the P1 promoter becomes nearly twofold stronger in vitro due to the presence of adenine tracks. These adenine residues occur periodically at –84.5, –74 and –63 positions.
- The adenine tracks bend the DNA at the P1 region. It is the process where the curvature of DNA influence the binding of RNA polymerase.
- RNA polymerase binds more efficiently to the bent face of P1. The bent structure helps the polymerase in forming a stable interaction.
- This DNA bending may produce a caging effect, which favour the formation of an RNA polymerase–promoter complex. This complex is better suited for transcription initiation at P1.
- Therefore, the adenine tracks act as intrinsic structural elements that increase P1 strength without involvement of any regulatory proteins.
Control of P2 by UTP
- It is observed that transcription from the P2 promoter becomes very low when the UTP level is high. When UTP is low, the activity of P2 increase both in vivo and in vitro.
- UTP directly influence the promoter clearing step of RNA polymerase at P2. It is the stage where the enzyme shift from initiation to elongation.
- At the P2 promoter the clearing is poor. RNA polymerase produce many abortive RNA fragments, which is different from P1 where clearing occur more smoothly.
- When UTP concentration is high, the enzyme also forms pseudo-templated oligomers pppAUn (n=2 to >20). This occurs because the polymerase stutters while adding uridines at positions 3–5 of the P2 transcript.
- At low UTP concentration, the polymerase clears the promoter more efficiently. It then produce regular gal RNA according to the template sequence.
- The regulation by UTP may be linked to the role of UTP in forming UDP sugars in the galactose pathway. It is the pathway that generate UDP-galactose and UDP-glucose.
- When UTP is high, the levels of UDP-galactose and UDP-glucose also increase. These sugars then inhibit the synthesis of their producing enzymes such as galactose-1-phosphate uridyltransferase and UDP-galactose epimerase.
- Thus, the control of P2 by UTP act as a feedback mechanism that coordinate nucleotide supply with galactose metabolism.
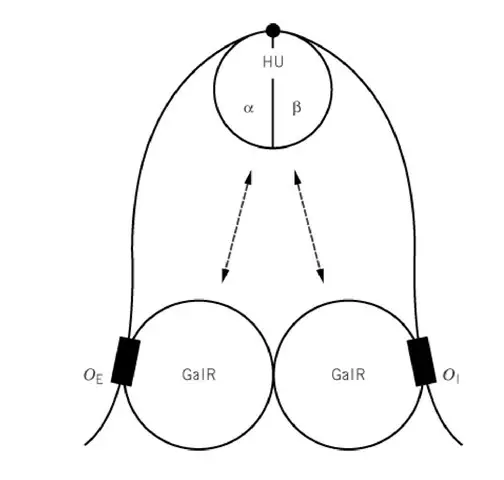
Repression by GalR and HU: DNA Looping
- GalR and HU repress transcription from P1 and P2 in a coordinated way. It is the negative control system that keep the promoters at low activity.
- The repression needs GalR binding to two operator sites. These are the external operator Oe and the internal operator Oi, each having 16-bp dyad-symmetric sequences.
- Oe is located at –60.5 upstream of the promoters and Oi is positioned at +53.5 inside the galE coding region.
- When GalR is bound at both operators, and when HU is present, the two GalR molecules interact. It is the process where a DNA loop is created over the promoter region.
- The DNA looping alters the structure of P1 and P2. The promoters become resistant to RNA polymerase caging and transcription initiation is suppressed.
- The combined complex of GalR, HU and operator DNA is called the Gal repressosome. It is the nucleoprotein assembly responsible for repression.
- GalR alone can bind the operators, but GalR alone cannot form the loop. HU is essential. Other histone-like proteins cannot replace HU.
- HU is a heterodimer (HUα and HUβ). A single HU molecule binds and bends the DNA at a critical architectural point that make the loop possible.
- Structural modelling show that GalR forms a V-shaped tetramer when bridging Oe and Oi. The resulting loop is antiparallel and cause DNA undertwisting.
- GalR and HU bind cooperatively. HU binding require GalR at both operators, and GalR binding is increased by HU. GalR helps to position HU at the correct DNA site.
- The GalR–HU interaction may be temporary and is not retained in the final repressosome, but it facilitate the assembly of the looped complex.
- Since HU binding depends on GalR, the repression complex becomes responsive to the inducer D-galactose. The inducer relieves repression by affecting GalR.
- This method allow DNA that was condensed and resistant to RNA polymerase to become open again when the proper signals appear.
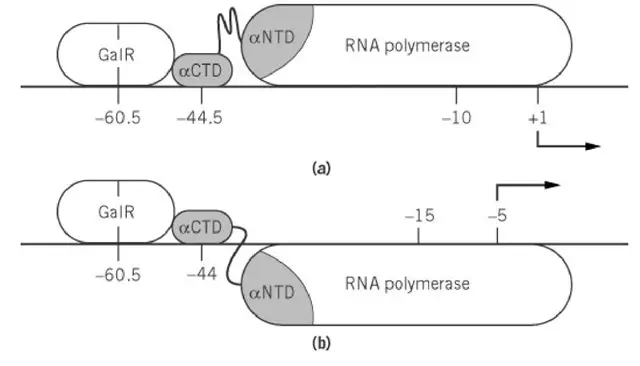
Regulation in the Absence of DNA Looping: Interaction between GalR and RNA Polymerase
- When DNA looping does not occur, such as when HU is absent, GalR binding only at Oe gives a dual control effect. It repress P1 by about four to five fold and it activates P2 nearly twofold.
- The occupancy of Oi does not change this behaviour. P1 repression and P2 activation continue independently of Oi.
- Each promoter respond separately. Even if both promoters is modified, GalR still gives its specific effect on whichever promoter is being examined.
- The activation at P2 or repression at P1 is not a fixed promoter property. It can be reversed when the angular orientation of the promoters to Oe is changed by inserting a 5-bp segment (half turn of DNA).
- For correct activation or repression, a specific GalR–RNA polymerase–DNA complex must be formed at each promoter.
- GalR acts directly on RNA polymerase. It is the process where the GalR protein contact the polymerase bound at the promoter.
- Activation of P2:
- GalR bound at –60.5 (–55.5 relative to P2) functions similar to many activators that bind at –60.
- The DNA-bound GalR interacts with the αCTD (α-subunit C-terminal domain) of RNA polymerase.
- The αCTD binds near 40 bp upstream of P2, and GalR contact helps the formation of the open complex.
- Repression of P1:
- When GalR binds at –60.5 relative to P1, it contact the αCTD bound 45 bp upstream of P1.
- This interaction reduce the isomerization of the RNA polymerase complex at P1 and transcription initiation becomes suppressed.
- It is still not known how the same GalR–polymerase interaction give activation at P2 and repression at P1. The conditions that decide this dual behaviour in cells without looping also remain unclear.
- Thus, in absence of DNA looping, the regulation depends entirely on the orientation, distance, and direct physical contact between GalR and the promoter-bound RNA polymerase.
Gal Isorepressor
- GalS is an isorepressor that also regulate the gal operon. It is related to GalR and act on similar operator sites.
- It does not form DNA looping. The repression or activation occur without the looping mechanism that needs HU.
- GalS binds to Oe and give the same pattern of control as GalR. It repress P1 and it stimulate P2, although the effect is weaker.
- GalR and GalS together control several other operons. These operons include the genes for galactose transport systems of both high and low affinity.
- The strength of control vary in different operons. It is the process that help coordinate galactose metabolism with the transport of the sugar in different conditions.
- GalS therefore act as a secondary regulator that fine-tune the expression of gal genes over a wide range of galactose availability.
Properties of GalR and GalS
- The amino acid sequences of GalR and GalS are highly similar. It is about 85% similarity, indicating that both proteins share common structural features.
- Both proteins are believed to contain two domains joined by a flexible hinge. This idea comes from their similarity to other members of the GalR–LacI family that have known structures.
- In the amino-terminal domain of each subunit, a helix–turn–helix motif is present. It is the region that recognises one half of the dyad-symmetric operator sequences at Oe and Oi.
- The carboxy-terminal domains contain the inducer-binding sites. These sites were identified from the study of noninducible mutants (galRs) and by structural modelling.
- The mechanism by which the inducer relieves repression at P1 under non-looping conditions has been deeply studied. It is the process where inducer binding change the activity of GalR at the promoter.
- Although inducer binding can cause GalR to detach from the operator, derepression of P1 can still occur even when GalR–inducer complex remains bound at Oe.
- This shows that removal of the repressor from DNA is not required for transcription. Instead, the OE-bound GalR normally interacts with RNA polymerase and block the formation of the open complex at P1.
- The inducer reduce this inhibitory contact allosterically. It weaken the protein–protein interaction without removing the repressor from the DNA.
- Thus GalR and GalS use a flexible, dual-domain structure to recognise operators and respond to inducer sugars by altering their contact with RNA polymerase.
Regulation by cAMP*CRP
- The cAMP*CRP complex is a global regulator. It gives opposite effects on the two gal promoters. It stimulate P1 about three- to fourfold, and it repress P2 nearly tenfold.
- This regulatory pattern is different from the lac promoter system. In lac, cAMP*CRP binds around –61.5 and activate transcription by contacting the αCTD of RNA polymerase.
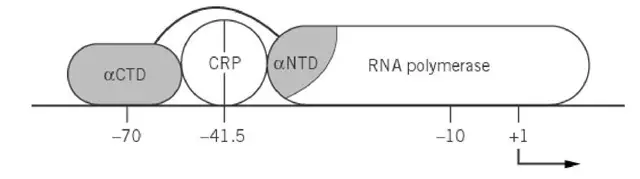
- In the gal operon the complex binds at position –41.5. This different binding position is responsible for the dual control on P1 and P2.
- P2 repression is believed to occur because cAMP*CRP blocks RNA polymerase binding at the overlapping –35 region. This prevent polymerase from initiating transcription at P2.
- Activation at P1 is achieved through a different mechanism. cAMP*CRP increase RNA polymerase binding (closed-complex formation) and also increase the isomerization step.
- A specific region (region 1) of the promoter-distal subunit of cAMPCRP contact helix 1 of the αCTD. This helps the αCTD to move away from the αNTD so that it can bind upstream of the cAMPCRP site.
- The promoter-proximal subunit interacts with a different part of the αNTD through another region (region 2). This promote the isomerization step.
- Thus the same regulator interacts with RNA polymerase at two different points. One interaction help polymerase binding, and the other stimulate the transition to open complex.
- These dual effects allow the operon to be expressed mainly from P1 when cAMP is abundant, while in cAMP-deficient conditions (like glucose-grown cells), transcription shift toward P2.
- Lewis DE, Adhya S. Molecular Mechanisms of Transcription Initiation at gal Promoters and their Multi-Level Regulation by GalR, CRP and DNA Loop. Biomolecules. 2015 Oct 16;5(4):2782-807. doi: 10.3390/biom5042782. PMID: 26501343; PMCID: PMC4693257.
- Semsey, S., Virnik, K., & Adhya, S. (2006). Three-stage Regulation of the Amphibolic gal Operon: From Repressosome to GalR-free DNA. Journal of Molecular Biology, 358(2), 355–363. doi:10.1016/j.jmb.2006.02.022
- Irani, M. H., Orosz, L., & Adhya, S. (1983). A control element within a structural gene: The gal operon of Escherichia coli. Cell, 32(3), 783–788. doi:10.1016/0092-8674(83)90064-8
- https://en.wikipedia.org/wiki/Gal_operon
- https://www.slideshare.net/Divyapeddapalyam/galactose-operon-slide-share
- http://what-when-how.com/molecular-biology/gal-operon-molecular-biology/
Greetings from Idaho! I’m bored to death at work so I decided
to browse your blog on my iphone during lunch break. I really like the info you present here and can’t
wait to take a look when I get home. I’m surprised
at how quick your blog loaded on my phone .. I’m not even using WIFI,
just 3G .. Anyways, fantastic site!
What’s up, its fastidious post on the topic of media print, we all know media is a fantastic
source of information.
Keep on writing, great job!
Thank You Very much @tanya
You actually make it appear so easy along with your presentation however I find this
matter to be really something which I feel I might never understand.
It sort of feels too complex and extremely vast for me.
I am having a look ahead to your subsequent publish, I’ll attempt to get
the grasp of it!
Hello there, I found your site by way of Google while looking for
a related subject, your web site came up, it appears great.
I’ve bookmarked it in my google bookmarks.
Hi there, just changed into alert to your blog thru Google, and located that
it’s really informative. I am gonna watch out for brussels.
I will appreciate for those who continue this in future.
Numerous other folks will be benefited from your writing.
Cheers!
Awesome website you have here but I was wondering if you knew of any forums that cover the same topics talked about here?
I’d really love to be a part of group where I can get advice from other experienced individuals that share the same interest.
If you have any recommendations, please let me know. Kudos!