What is gab operon?
- The gab operon is in charge of turning -aminobutyrate (GABA) into succinate.
- The gab operon is made up of three structural genes: gabD, gabT, and gabP. These genes code for an enzyme called succinate semialdehyde dehydrogenase, an enzyme called GABA transaminase, and a protein called GABA permease.
- Downstream of the operon is a regulatory gene called csiR. It codes for a putative transcriptional repressor and is turned on when nitrogen is scarce.
- The gab operon has been studied in Escherichia coli, and enzymes from Saccharomyces cerevisiae, rats, and humans have been found to have a lot in common with those from Escherichia coli.
- When there isn’t enough nitrogen, the gab genes are turned on.
- These genes make enzymes that change GABA into succinate. Succinate then goes into the TCA cycle and is used as a source of energy.
- The gab operon is also known to help maintain polyamine homeostasis when there isn’t enough nitrogen for growth and to keep glutamate levels high when there is stress.
- GABA transaminase is an enzyme that changes GABA and 2-oxoglutarate into succinate semialdehyde and glutamate. It is made by the gabT gene.
- Then, succinate semialdehyde is turned into succinate by an enzyme called succinate semialdehyde dehydrogenase, which is made by the gabP gene. Succinate then enters the TCA cycle as an energy source.
- During growth when nitrogen is scarce, the gab operon helps keep polyamines like putrescine in balance. It is also known to keep its glutamate levels high even when it is under stress.
Structure of gab operon
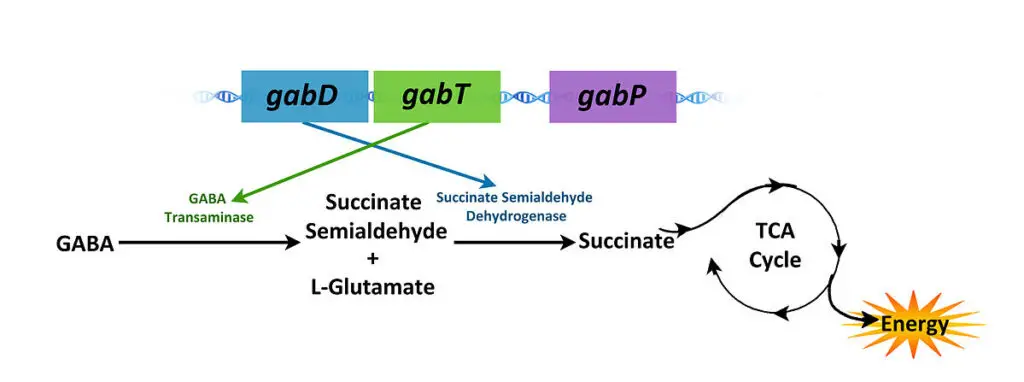
There are three structural genes that make up the gab operon:
- gabT : The gabT gene encodes a GABA transaminase that generates succinic semialdehyde.
- gabD: encodes a NADP-dependent succinic semialdehyde dehydrogenase, which oxidises succinic semialdehyde to succinate.
- gabP: encodes a GABA-specific permease.
Regulation of gab operon
Differential Regulation of Promoters
The expression of the genes in the operon is controlled by three promoters that work in different ways. Two of these promoters are controlled by the sigma factor S that is encoded by RpoS.
- csiDp: csiDp is dependent on S and only turns on when there isn’t enough carbon. This is because cAMP-CRP is the only thing that can turn on S-containing RNA polymerase at the csiD promoter.
- gabDp1: depends on S and is caused by a number of stresses.
- gabDp2: gabDp2 is controlled by regulatory proteins called Nac (Nitrogen Assimilation Control) when there is not enough nitrogen.
Mechanism of gab operon Regulation
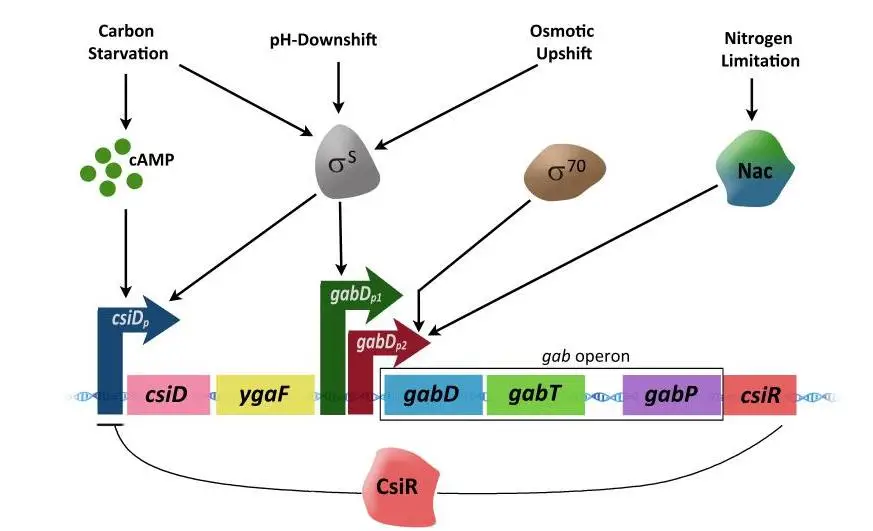
Activation of gab operon
- The carbon starvation-induced gene (csiD), the ygaF gene, and the gab genes can’t be turned on or off without the csiD promoter (csiDp).
- The csiDp is only turned on when there isn’t enough carbon in the cell and the cell is in a stationary phase where cAMP levels are high.
- When cAMP binds to the cAMP receptor protein (CRP), CRP tightly binds to a specific DNA site in the csiDp promoter. This turns on the transcription of genes that are further down the chain from the promoter.
- The gabDp1 controls the gabDTP region in a different way.
- In addition to starvation and stationary phase, S can also cause hyperosmotic and acidic shifts, which turn on gabDp1.
- The gabDp2 promoter, on the other hand, depends on 70 and is turned on when there isn’t enough nitrogen.
- In situations where there isn’t enough nitrogen, the nitrogen regulator Nac binds to a site just upstream of the promoter that tells the gab genes to be turned on.
- When the gab genes are turned on, they make enzymes that break down GABA into succinate.
Repression
- When nitrogen is present, it turns on the csiR gene, which is located after the gabP gene.
- The csiR gene makes a protein that stops the csiD-ygaF-gab operon from being copied. This turns off the GABA degradation pathway.
GABA degradation pathways are found in almost all eukaryotic organisms and are done by enzymes that work the same way. Even though E. coli mostly uses GABA as an alternative source of energy through GABA degradation pathways, higher eukaryotic organisms use GABA as an inhibitory neurotransmitter and as a muscle tone regulator. In eukaryotes, GABA is broken down by pathways called GABA degradation pathways.
References
- Joloba, Moses & Clemmer, Katy & Sledjeski, Darren & Rather, Philip. (2005). Activation of the gab Operon in an RpoS-Dependent Manner by Mutations That Truncate the Inner Core of Lipopolysaccharide in Escherichia coli. Journal of bacteriology. 186. 8542-6. 10.1128/JB.186.24.8542-8546.2004.
- https://dbpedia.org/page/Gab_operon
- https://en.wikipedia.org/wiki/Gab_operon
- https://www.wikiwand.com/en/Gab_operon