What are exons?
- An exon is any part of a gene that forms a part of the final mature RNA after introns have been removed by RNA splicing.
- Exons refer to both the DNA sequence within a gene and the corresponding sequence in RNA transcripts.
- In RNA splicing, introns are removed, and exons are covalently joined to generate mature RNA.
- The term “exon” was coined by Walter Gilbert in 1978 and derives from the expressed region of a gene.
- Initially, exons were defined for protein-coding transcripts that are spliced before translation.
- Exons also include sequences removed from rRNA, tRNA, and other non-coding RNA molecules.
- Exons encode portions of the final product of a gene, such as proteins, in eukaryotic genes.
- Exons are exclusively present in eukaryotic genes.
- Exons are the coding regions of a gene and are important segments.
- Introns, non-coding regions, interspace exons in a gene.
- Splicing, a post-transcriptional process, removes introns and joins exons.
- Exons contain the information required to encode proteins.
- Genes in eukaryotes consist of coding exons interspersed with non-coding introns.
- Introns are removed to create a functional messenger RNA (mRNA) for protein translation.
The history of the term “exon” dates back to 1978 when Walter Gilbert, an American biochemist, introduced the concept. Gilbert proposed a new perspective on the structure of genes, suggesting that the traditional idea of a cistron should be replaced by a transcription unit consisting of regions that would be lost from the mature messenger RNA (mRNA), which he referred to as introns, alternating with regions that would be expressed, which he termed exons.
Initially, this definition of exons was specifically applied to protein-coding transcripts that undergo splicing before translation. The focus was on the regions of genes that would be transcribed into mature mRNA and eventually translated into proteins.
Over time, the definition of exons expanded to include other types of RNA molecules. Sequences removed from ribosomal RNA (rRNA), transfer RNA (tRNA), and other non-coding RNA (ncRNA) were also referred to as exons. This broadening of the term encompassed a wider range of RNA molecules originating from different parts of the genome.
Furthermore, the concept of exons evolved to include trans-splicing, a process where RNA molecules from different genomic regions are ligated together. In this context, exons referred to the segments of RNA molecules originating from different parts of the genome that were joined through trans-splicing.
Overall, the term “exon” has undergone expansion and refinement since its inception, starting with its application to protein-coding transcripts and later incorporating other types of RNA molecules and trans-splicing events. This evolving understanding of exons has contributed to our knowledge of gene structure and RNA processing in various biological contexts.
Exon Definition
Exon: A coding region of a gene that contains the information necessary to produce a protein or is incorporated into RNA structure.
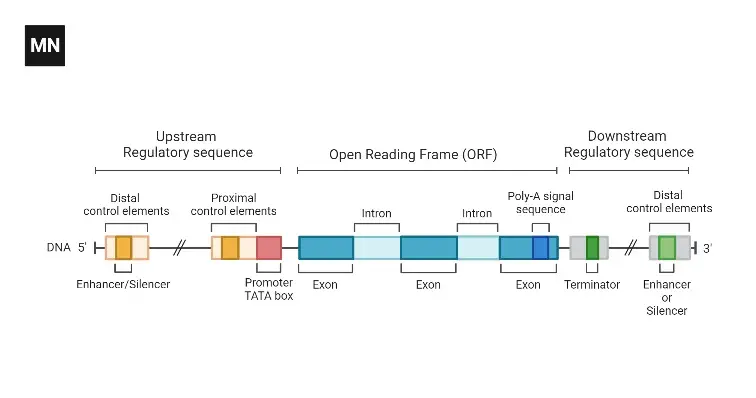
Structure of Exon
- Exons are nucleotide sequences in DNA or RNA that are expressed.
- Exons are conserved in both DNA and mature RNA.
- Exon structure includes both the coding sequence and the untranslated regions (UTRs) from the 5′ and 3′ ends.
- In protein-coding genes, the first exon often contains both the 5′-UTR and the initial part of the coding sequence, but exons consisting solely of UTR regions or 3′-UTR regions can also occur.
- Non-coding RNA transcripts can also contain exons and introns.
- Alternative splicing can result in different mature mRNA molecules originating from the same gene having varying sets of exons, as different introns in the pre-mRNA can be removed.
- Exonization refers to the creation of new exons through mutations in introns.
- Exons are composed of stretches of DNA that will be translated into amino acids and proteins.
- In eukaryotic organisms, exons can be contiguous within a gene or separated by introns in a discontinuous gene.
- During transcription, the gene is transcribed into pre-mRNA, which contains both introns and exons.
- The pre-mRNA undergoes processing, where introns are spliced out, resulting in mature mRNA.
- Mature mRNA consists of exons and short untranslated regions (UTRs) at both ends.
- The exons form the final reading frame, which consists of nucleotides arranged in triplets.
- The reading frame begins with a start codon (usually AUG) and ends with a termination codon.
- The nucleotides in the reading frame are arranged in triplets, with each triplet coding for an amino acid.
What is Splicing?
Splicing is a crucial process in gene expression that involves the removal of introns, the non-coding regions, from the primary mRNA transcript and the joining together of exons to form a mature mRNA molecule. Here are some key points about splicing:
- Formation of mature mRNA: Splicing transforms the initial pre-mRNA, which contains both exons and introns, into a mature mRNA molecule that contains only the coding exons. This mature mRNA serves as the template for protein synthesis.
- Occurs before translation: Splicing takes place prior to the process of translation, which is the synthesis of proteins. It ensures that the correct genetic information is preserved in the mRNA for proper protein production.
- Importance for protein coding: Splicing is essential for the accurate coding of proteins. Without splicing, the protein-coding regions would be interrupted by non-coding introns, leading to the production of non-functional or truncated proteins.
- Role in gene expression regulation: Splicing plays a significant role in regulating gene expression. Alternative splicing allows for the production of different mRNA isoforms from a single gene, leading to the synthesis of various protein variants with distinct functions. This contributes to the complexity and diversity of gene expression.
- Factors influencing splicing: Splicing can vary based on the type of organism, the RNA or intron structure, and the presence of specific catalysts. Different organisms and cell types may exhibit different splicing patterns and mechanisms.
- Splicing methods: Several mechanisms and pathways for splicing have been identified, including the major spliceosome pathway and alternative splicing pathways. These mechanisms involve the precise recognition and removal of introns and the precise joining of exons to generate the mature mRNA.
Splicing Process
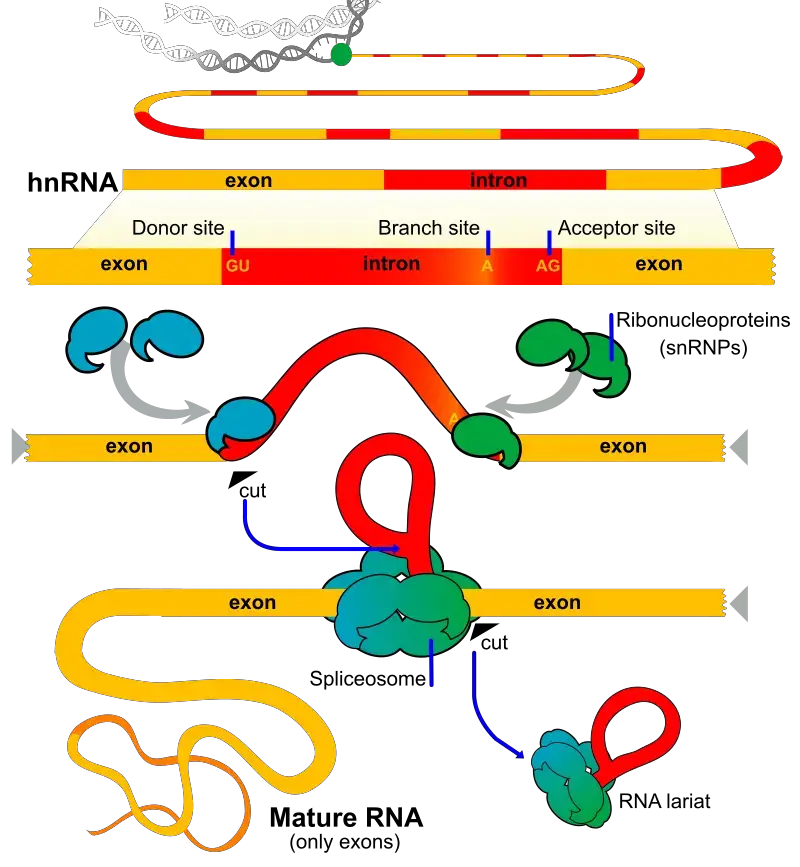
The process of splicing involves the recognition and removal of introns from pre-mRNA to generate mature mRNA. Here are the key steps involved in splicing:
- Conserved sequences: The pre-mRNA contains exon-intron junctions with conserved sequences. The 5′ end junction has a GU sequence, known as the 5′ splice site, while the 3′ end junction has an AG sequence, called the 3′ splice site.
- Branch point site: Upstream of the 3′ splice site, there is an invariant site located 15-45 nucleotides away. This site is rich in adenine (A) bases and is known as the branch point or branch site.
- Recognition of splice sites: The spliceosome, a complex composed of RNA and protein components, recognizes these conserved sequences in the pre-mRNA at the respective splice sites. The spliceosome is composed of small nuclear ribonucleoproteins (snRNPs), which consist of small nuclear RNA (snRNA) and associated proteins.
- Composition of spliceosome: The key snRNPs involved in spliceosome formation are U1, U2, U5, and the U4-U6 complex. U4 and U6 are bound together as a single entity within the spliceosome.
- Spliceosome assembly: The spliceosome assembles onto the pre-mRNA, bringing together the snRNPs and other associated proteins at the splice sites. This complex formation allows for precise recognition and binding of the splice sites.
- Spliceosome catalysis: The spliceosome catalyzes two transesterification reactions. The first reaction involves cleavage at the 5′ splice site, resulting in the formation of a free 5′ end of the intron and an intron lariat structure linked to the branch point. In the second reaction, the intron is released, and the adjacent exons are joined together, forming a continuous mRNA molecule.
- Exon ligation: The exons are ligated together by the spliceosome, and the intron is released as a lariat structure. The resulting mature mRNA contains only the coding exons and is ready for translation into a protein.
Overall, the process of splicing involves the recognition of conserved sequences at the exon-intron junctions, the assembly of the spliceosome, catalysis of the splicing reactions, and the ligation of exons to generate mature mRNA. This precise and regulated process ensures the accurate removal of introns and the proper joining of exons, leading to the production of functional mRNA for protein synthesis.
Splicing Pathway
The splicing pathway involves a series of steps facilitated by small nuclear ribonucleoproteins (snRNPs) to remove introns and join exons in pre-mRNA. Here is an overview of the splicing pathway:
- U1 and U2 binding: U1 snRNP binds to the 5′ splice site, and U2 snRNP binds to the invariant site or the branch point within the intron.
- Binding of additional snRNPs: Other snRNPs, including U5 and U4-U6, bind to the intron region, leading to the assembly of the spliceosome complex.
- Spliceosome assembly: The spliceosome complex forms by the binding of snRNPs and associated proteins. This complex brings the intron and exon sequences into close proximity.
- Looping and intron removal: The spliceosome loops out the intron, bringing the two ends of the intron close to each other.
- Release and rearrangement of snRNPs: U1 and U4 snRNPs are released from the spliceosome, while U6 snRNP is bound to both the 5′ splice site and U2 snRNP.
- Cleavage and ligation: The 5′ end of the intron is cleaved, and the cleaved end attaches to the branch point, which is rich in adenine (A). Subsequently, the 3′ end of the intron is cleaved, resulting in the release of the intron in a loop-like structure called a lariat.
- Exon joining: The two adjacent exons are ligated together, forming a continuous mature mRNA molecule.
- Continued splicing: The snRNPs that were involved in the splicing of one intron are released and can be reused for splicing of other introns within the pre-mRNA. The splicing process continues until all the introns have been removed.
- Intron degradation: The released intron, in its lariat form, is further degraded by enzymes, and its components can be recycled for other cellular processes.
By following this splicing pathway, the spliceosome complex ensures the precise removal of introns and the accurate joining of exons, resulting in the generation of mature mRNA that can be translated into proteins.
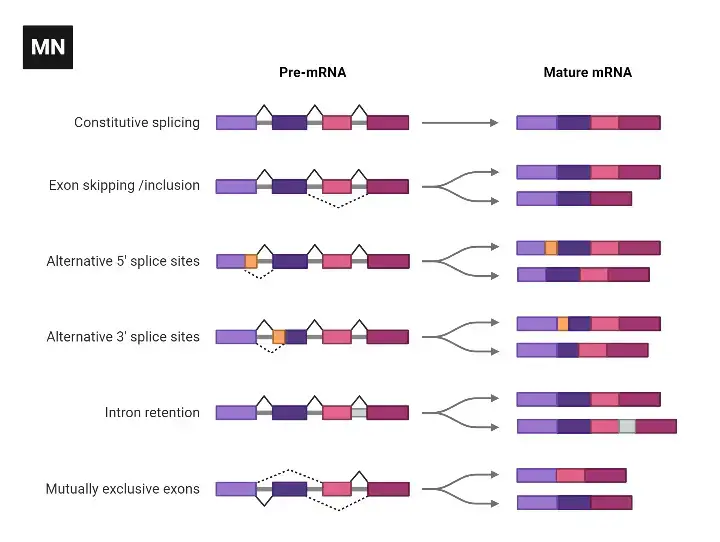
Alternative Splicing
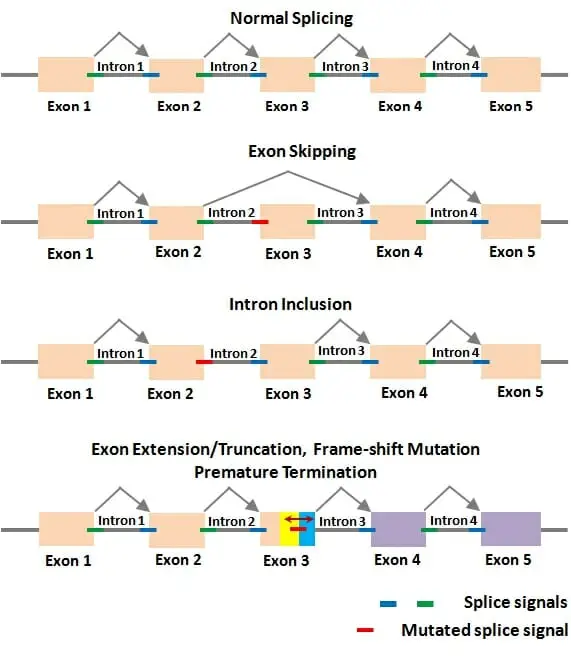
Alternative splicing is a mechanism that allows for the production of different mRNA isoforms by selectively joining different exons together during RNA processing. This process leads to the generation of multiple protein variants from a single gene. Here are the main types of alternative splicing:
- Consecutive splicing: In this type of alternative splicing, consecutive introns are spliced out, and the adjacent exons are joined together. This results in the inclusion of specific exons and the exclusion of others, creating mRNA variants with different exon compositions.
- Exon skipping: In exon skipping, certain exons, along with their adjacent exons, are excised from the pre-mRNA before translation. This results in the exclusion of specific exons from the final mRNA, leading to the production of protein isoforms lacking those exons.
- Alternative 5′ splice site or 3′ splice site: Alternative splicing can also occur by utilizing alternative 5′ or 3′ splice sites. This means that different exons can be joined together at alternative splice sites, resulting in mRNA isoforms with different exon inclusion patterns.
- Intron retention: In some cases, certain introns are retained in the mature mRNA instead of being spliced out. This leads to the inclusion of intronic sequences within the mRNA and can have functional consequences for the resulting protein isoform.
The phenomenon of alternative splicing greatly increases the diversity of proteins that can be generated from a limited number of genes. By selectively including or excluding exons, alternative splicing can modify the coding sequence and, consequently, the structure, function, and regulation of proteins. It plays a critical role in tissue-specific gene expression, developmental processes, and the generation of protein diversity in complex organisms.
Function of Exon
- Exons encode proteins: Exons are segments of coding DNA that carry the information necessary to produce proteins. Different exons within a gene can encode different domains of a protein.
- Formation of protein domains: Exons may encode protein domains individually or can be spliced together to form functional protein domains. This splicing of exons allows for the creation of diverse protein structures and functions.
- Molecular evolution through exon shuffling: The presence of exons and introns enables molecular evolution through a process called exon shuffling. Exon shuffling occurs when exons are exchanged between sister chromosomes during recombination, leading to the formation of new genes with different combinations of exons.
- Alternative splicing: Exons also play a crucial role in alternative splicing, a process that allows for the production of multiple protein isoforms from a single gene. Alternative splicing involves different arrangements of exons when introns are removed during RNA processing. This can include the complete exclusion of an exon, partial inclusion of an exon, or even the inclusion of parts of introns.
- Generation of protein diversity: Alternative splicing enables the generation of multiple protein variants from a single gene, increasing protein diversity and functional complexity. Alternative splicing can occur in the same location to produce different isoforms with similar roles or in different cell or tissue types, expanding the range of protein functions.
- Implications in disease: Alternative splicing, as well as defects in alternative splicing, can contribute to various diseases. Conditions like alcoholism and cancer have been associated with abnormalities in alternative splicing, highlighting the importance of properly regulated exon usage in maintaining normal cellular function.
FAQ
What is an exon?
An exon is a segment of DNA or RNA that contains coding sequences and is involved in the production of mature RNA molecules.
What is the role of exons in gene expression?
Exons play a crucial role in gene expression by encoding the sequences that will be translated into proteins. They are responsible for the coding regions of a gene.
How are exons and introns different?
Exons are coding regions of a gene that contain information for protein synthesis, while introns are non-coding regions that are interspersed between exons.
How are exons and introns related to RNA splicing?
RNA splicing is the process by which introns are removed from pre-mRNA, and exons are joined together to form mature mRNA that can be translated into proteins.
Can exons code for different domains of a protein?
Yes, exons can code for different domains of a protein. Different exons can be spliced together in various combinations to generate protein isoforms with distinct functional domains.
What is alternative splicing, and how does it relate to exons?
Alternative splicing is a process where different combinations of exons are selectively joined together, resulting in the production of multiple mRNA isoforms from a single gene. Alternative splicing increases protein diversity.
How do exons contribute to protein diversity?
Exons contribute to protein diversity through alternative splicing. By including or excluding specific exons, different protein isoforms can be generated from a single gene, each with unique functional properties.
Are exons conserved across different species?
Exons generally exhibit higher conservation compared to introns. Coding sequences that are responsible for protein synthesis tend to be more conserved across different species due to their functional importance.
Can mutations occur in exons?
Yes, mutations can occur in exons. Mutations in exons can lead to changes in the amino acid sequence of proteins, potentially affecting protein function and leading to genetic disorders or diseases.
How do exons impact gene regulation and protein function?
Exons can influence gene regulation by containing regulatory elements such as enhancers or silencers. Additionally, different exons can produce protein isoforms with varying structures or functional domains, impacting protein function and cellular processes.
References
- https://www.genome.gov/genetics-glossary/Exon
- https://en.wikipedia.org/wiki/Exon
- https://biologydictionary.net/exon/
- https://www.news-medical.net/life-sciences/What-are-introns-and-exons.aspx
- https://www.biologyonline.com/dictionary/exon
- https://www.aatbio.com/resources/faq-frequently-asked-questions/What-is-exon-shuffling
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.