What are Enzymes?
- Enzymes, often referred to as the “biological catalysts,” play a pivotal role in the metabolic processes within living organisms. These protein biomolecules are responsible for accelerating various biochemical reactions, ensuring that life-sustaining processes occur at a pace conducive to life.
- Derived from the term “in yeast,” enzymes were initially identified in the context of their role in the production of ethyl alcohol and carbon dioxide through the enzyme zymase in yeast. These biological catalysts are distinct from their chemical counterparts in that they are proteins, making them susceptible to damage and inactivation. Their specificity is noteworthy, as each enzyme typically acts on a particular substrate for a specific reaction.
- Enzymes can be succinctly defined as biological polymers that facilitate biochemical reactions. They are integral to numerous metabolic processes, with each enzyme tailored to catalyze specific reactions. The initial phase of any metabolic process hinges on enzymes, which interact with a molecule known as the substrate. Through this interaction, enzymes transform substrates into products, a process fundamental to life.
- The intricate nature of enzymes extends to their composition. While the majority are protein-based, a class of RNA catalysts, termed ribozymes, also exists. These ribozymes are ribonucleic acid molecules capable of catalyzing reactions within their own bonds or among other RNAs. It’s fascinating to note that enzymes are omnipresent, found in every tissue and fluid within the body. Their role is not just limited to metabolic pathways; enzymes in the plasma membrane respond to cellular signals, while those in the circulatory system are pivotal for blood clotting.
- Therefore, understanding enzymes is crucial due to their role in life processes. Their specificity arises from their unique three-dimensional structure. Besides accelerating reactions by lowering the activation energy, enzymes ensure that reactions occur millions of times faster than they would otherwise. For instance, the enzyme orotidine 5′-phosphate decarboxylase facilitates a reaction in milliseconds that would naturally take millions of years.
- Then, it’s essential to recognize that enzymes, like all catalysts, do not alter the equilibrium of a reaction. Their specificity sets them apart from other catalysts. External molecules can influence enzyme activity, with inhibitors decreasing it and activators enhancing it. Many drugs and poisons function as enzyme inhibitors. Enzymes also exhibit sensitivity to changes in temperature and pH, with excessive heat leading to denaturation and loss of their catalytic properties.
- Besides their biological significance, enzymes have commercial applications. They are employed in antibiotic synthesis and are integral to certain household products. For instance, enzymes in detergents help break down stains, while those in meat tenderizers make meat more palatable by breaking down proteins.
- In conclusion, enzymes, with their proteinaceous nature, are the linchpins of life processes. Their ability to catalyze specific reactions, combined with their intricate structure and function, underscores their importance in both biological and commercial realms. Their study, termed enzymology, continues to shed light on their myriad roles and mechanisms, emphasizing their significance in sustaining life.
Definition of Enzymes
Enzymes are protein molecules that act as biological catalysts, accelerating chemical reactions within living organisms without being consumed in the process.
Characteristics Features of Enzymes
Enzymes are biological molecules with distinct characteristics and features that enable them to play crucial roles in various biochemical processes. Here are the key characteristics and features of enzymes:
- Catalytic Activity: Enzymes are highly efficient catalysts that accelerate chemical reactions in living organisms. They facilitate reactions by lowering the activation energy required for the reaction to occur, thus increasing the reaction rate.
- Specificity: Enzymes exhibit remarkable specificity. Each enzyme is highly selective for a particular substrate or group of structurally related substrates. This specificity is essential for the precise regulation of biochemical pathways.
- Protein Nature: Enzymes are primarily composed of protein molecules. The protein component of an enzyme is known as the apoenzyme, and it provides the structural framework for the enzyme’s function.
- Active Site: Enzymes have an active site, which is a specific region on the enzyme’s surface where the substrate binds. The active site is often a pocket or cleft on the enzyme’s protein structure.
- Lock-and-Key Model: The interaction between enzymes and substrates is often described by the lock-and-key model. In this model, the active site of the enzyme has a shape that precisely matches the shape of the substrate, ensuring a snug fit.
- Induced Fit: Another model, known as the induced fit model, suggests that the enzyme’s active site can change its shape slightly to accommodate the substrate, leading to optimal binding.
- Reusability: Enzymes are not consumed or altered during the reaction. After catalyzing a reaction, they are released and can be used again for subsequent reactions, making them highly efficient.
- pH and Temperature Sensitivity: Enzymes have specific pH and temperature ranges at which they function optimally. Deviations from these ranges can lead to decreased enzyme activity or denaturation.
- Cofactors and Coenzymes: Many enzymes require cofactors or coenzymes to function effectively. Cofactors can be inorganic ions, while coenzymes are organic molecules. They assist enzymes in catalyzing reactions.
- Regulation: Enzyme activity is often regulated to maintain metabolic homeostasis. Regulation can occur through allosteric modulation, competitive or non-competitive inhibition, and post-translational modifications.
- Enzyme Naming: Enzymes are typically named based on their substrates and the type of reaction they catalyze, often ending with the suffix “-ase.” For example, “lipase” catalyzes lipid hydrolysis.
- Enzyme Kinetics: Enzyme activity follows kinetic principles, such as Michaelis-Menten kinetics, which describes the relationship between substrate concentration and reaction rate.
- Diversity: Enzymes are involved in a wide range of biological processes, including metabolism, DNA replication, digestion, and cell signaling. They come in various types and perform diverse functions.
- Enzyme Classes: Enzymes are classified into several classes based on the type of reaction they catalyze. These classes include oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases.
- Essential for Life: Enzymes are essential for the survival and functioning of all living organisms. Without enzymes, many biological reactions would occur too slowly to sustain life.
Nomenclature of Enzymes
Enzymes, the biological catalysts, have a specific naming system to ensure clarity and precision in their identification. This nomenclature is twofold: the recommended name and the systematic name.
- Recommended Name The recommended name is the shorter version, primarily used for daily references. Typically, these names end with the suffix “-ase.” This suffix is either attached to the substrate the enzyme acts upon, like in “glucosidase” or “urease,” or it describes the action the enzyme performs, such as “lactate dehydrogenase” or “adenylyl cyclase.” However, it’s essential to note that some enzymes retain their original names, which might not indicate the enzymatic reaction they are associated with. Examples of these are “trypsin” and “pepsin.”
- Systematic Name The systematic naming system is more comprehensive. Here, enzymes are categorized into six primary classes. Within these classes, enzymes are further divided into numerous subgroups. In this system, the “-ase” suffix is attached to a detailed description of the chemical reaction the enzyme catalyzes. This description includes the names of all substrates involved. For instance, “lactate:NAD+ oxidoreductase” is a systematic name. Besides, each enzyme is given a unique classification number for precise identification. For example, the classification number for lactate:NAD+ oxidoreductase is 1.1.1.27. Although systematic names provide clarity and are unambiguous, they can be quite lengthy and might not be suitable for general use.
Furthermore, it’s crucial to understand some potentially confusing aspects of enzyme nomenclature. For instance, “synthetase” indicates an enzyme that requires ATP, while “synthase” does not. “Phosphatase” uses water to remove a phosphoryl group, whereas “phosphorylase” uses Pi to break a bond, producing a phosphorylated product. “Dehydrogenase” involves NAD+/FAD as an electron acceptor in a redox reaction, “oxidase” has O2 as the acceptor without incorporating oxygen atoms into the substrate, and “oxygenase” incorporates one or both oxygen atoms.
Therefore, understanding the nomenclature of enzymes is essential for accurate communication in the field of biochemistry. Besides providing clarity, it ensures that enzymes are identified without any ambiguity. Then, with the consistent use of technical vocabulary and emphasis on functions, the nomenclature becomes a valuable tool for professionals and researchers in the field.
Properties of Enzymes
Enzymes, the biological catalysts, play a pivotal role in facilitating chemical reactions within organisms. These protein catalysts expedite the rate of chemical reactions without undergoing any change themselves. It’s noteworthy that while most enzymes are proteins, certain RNAs, termed ribozymes, also exhibit enzymatic activities, primarily catalyzing the synthesis and cleavage of phosphodiester bonds. However, ribozymes are less prevalent compared to protein catalysts.
- Active Sites Central to an enzyme’s function is its active site, a specialized pocket or cleft formed due to the protein’s folding. This site houses specific amino acid side chains that are crucial for substrate binding and the subsequent catalytic action. When a substrate binds to the enzyme, it forms an enzyme-substrate (ES) complex. This binding is believed to induce a conformational shift in the enzyme, a phenomenon described by the induced fit model. This ES complex then transforms into an enzyme-product (EP) complex, which eventually releases the enzyme and the product.
- Catalytic Efficiency The efficiency of enzyme-catalyzed reactions is remarkable, with these reactions occurring at a pace 10^3 to 10^8 times swifter than their uncatalyzed counterparts. The turnover number, also known as kcat, represents the number of substrate molecules an enzyme converts to product in a second. This number typically ranges between 10^2 and 10^4s^-1.
- Specificity Enzymes exhibit high specificity. They interact with specific substrates and facilitate distinct chemical reactions. The assortment of enzymes synthesized in a cell dictates the reactions that transpire within that cell.
- Holoenzymes, Apoenzymes, Cofactors, and Coenzymes Certain enzymes necessitate additional non-protein molecules for their activity. A holoenzyme denotes the active enzyme inclusive of its non-protein component. In contrast, an apoenzyme, devoid of this non-protein component, remains inactive. When this non-protein entity is a metal ion like Zn^2+ or Fe^2+, it’s termed a cofactor. If it’s an organic molecule, it’s labeled a coenzyme. Coenzymes that associate with the enzyme temporarily are known as cosubstrates. These cosubstrates, after dissociation, are altered, as seen with NAD+. Conversely, if a coenzyme remains permanently attached and reverts to its original form, it’s termed a prosthetic group, like FAD. Many coenzymes originate from vitamins, with NAD+ containing niacin and FAD containing riboflavin.
- Regulation Enzymatic activity is not static; it can be modulated based on the cell’s requirements. This regulation ensures that product formation aligns with the cell’s needs.
- Location within the Cell Enzymes are not randomly dispersed within a cell. Many are strategically positioned in specific organelles. This compartmentalization isolates the substrate or product from other reactions, offering a conducive environment for the reaction and systematically organizing the myriad enzymes in the cell into functional pathways.
Structure of Enzymes
Enzymes, the biological catalysts, play a pivotal role in various biochemical reactions within organisms. These proteins, characterized by their intricate structure, are composed of linear chains of amino acids that fold into a unique three-dimensional conformation. This article delves into the detailed structure of enzymes, emphasizing their functions and the significance of each structural component.
- Primary Structure – At the most basic level, enzymes consist of amino acid sequences linked by peptide bonds, forming a linear chain known as the primary structure. This sequence is not random; rather, it is dictated by the DNA sequence of the gene encoding the enzyme. Therefore, even a slight alteration in this sequence can drastically affect the enzyme’s function.
- Secondary Structure – Beyond the primary structure, amino acids within the chain interact with each other, primarily through hydrogen bonding. This interaction leads to the formation of the enzyme’s secondary structure, which can either be an α-helix or a β-sheet. In the α-helix structure, the protein chain coils around itself, while in the β-sheet, it folds back and forth.
- Tertiary Structure – The secondary structures further fold and interact, giving rise to the enzyme’s tertiary structure. This three-dimensional conformation is crucial for the enzyme’s functionality. It ensures that specific regions of the enzyme, vital for its catalytic activity, are correctly positioned.
- Active Site – Central to an enzyme’s function is its active site, a specialized region where substrates bind and undergo chemical transformation. The active site comprises the catalytic and binding sites, which work in tandem to facilitate the enzyme’s specific reaction. Only a minuscule portion of the enzyme, determined by its amino acid sequence, is involved in this catalytic activity.
Examples of Enzyme Structures
- Ribonuclease (RNase): This enzyme, involved in the hydrolysis of ribonucleic acids, consists of a single polypeptide chain of 124 amino acids. Approximately 25% of its structure is in the α-helix form, while the rest adopts the β-sheet conformation. The enzyme’s active site, crucial for its function, is located in a depression in the middle of the chain.
- Lysozyme: Found in various bodily secretions, lysozyme is composed of 129 amino acids. Its structure is predominantly α-helical, with about 12% being in the β-conformation. The enzyme’s active site, which binds to substrates, comprises six subsites.
- Chymotrypsin: This digestive enzyme, responsible for protein hydrolysis, is made up of three polypeptide chains. Its structure is characterized by antiparallel β pleated sheet regions interspersed with α-helical segments.
Structural Features of Enzymes
Enzymes exhibit a range of structural features that contribute to their function as biological catalysts. These structural characteristics enable enzymes to carry out their specific roles in biochemical reactions. Here are some key structural features of enzymes:
- Protein Composition: Enzymes are primarily composed of protein molecules. The protein component of an enzyme is called the apoenzyme. Proteins are made up of amino acids, and the sequence and arrangement of these amino acids determine the enzyme’s three-dimensional structure.
- Active Site: The active site is a crucial structural feature of enzymes. It is a specific region on the enzyme’s surface where the substrate binds and the catalytic reaction occurs. The active site often forms a pocket or cleft within the enzyme’s protein structure.
- Substrate Binding: Enzymes have a high degree of specificity for their substrates. The active site’s shape and chemical properties are complementary to those of the substrate, allowing for precise binding. This specificity ensures that enzymes catalyze specific reactions.
- Lock-and-Key Model: The lock-and-key model of enzyme-substrate interaction describes the precise fit between the substrate and the active site, much like a key fitting into a lock. This model emphasizes the importance of the active site’s specific shape.
- Induced Fit: In the induced fit model, the active site can undergo conformational changes upon substrate binding. The binding of the substrate induces structural adjustments in the enzyme, optimizing the interaction between the two.
- Cofactors: Some enzymes require cofactors to function effectively. Cofactors can be inorganic ions (e.g., metal ions like Mg²⁺) or organic molecules. Cofactors often assist in substrate binding or catalytic reactions.
- Coenzymes: Coenzymes are organic molecules that function as cofactors. They participate directly in the catalytic reactions by donating or accepting chemical groups. Common coenzymes include NAD⁺, NADP⁺, FAD, and coenzyme A.
- Quaternary Structure: Some enzymes are composed of multiple protein subunits that come together to form a quaternary structure. These subunits work cooperatively to create an active enzyme complex.
- pH Sensitivity: Enzyme activity is influenced by the pH of the surrounding environment. Many enzymes have an optimal pH range in which they function most effectively. Deviations from this pH range can lead to decreased enzyme activity.
- Temperature Sensitivity: Enzymes also have specific temperature ranges for optimal activity. Temperatures outside this range can result in denaturation, where the enzyme loses its structural integrity and activity.
- Allosteric Sites: Some enzymes have allosteric sites, which are distinct from the active site. Molecules binding to allosteric sites can modulate the enzyme’s activity, either enhancing or inhibiting it.
- Enzyme Kinetics: Enzyme activity follows kinetic principles, such as Michaelis-Menten kinetics. These principles describe the relationship between substrate concentration, reaction rate, and enzyme activity.
- Regulatory Sites: Enzymes can be regulated by various mechanisms, including competitive and non-competitive inhibition. Regulatory molecules can bind to specific sites on the enzyme to modulate its activity.
- Enzyme Classes: Enzymes are classified into different classes based on the type of reactions they catalyze. These classes include oxidoreductases, transferases, hydrolases, lyases, isomerases, and ligases.
- Diversity: Enzymes exhibit structural diversity to match the wide range of biochemical reactions in living organisms. Each enzyme’s structure is tailored to its specific function.
Types/Classification of Enzymes
The International Union of Biochemists (I U B) has systematically classified enzymes into six primary categories based on the type of reaction they catalyze. These categories are:
- Oxidoreductases: These enzymes facilitate oxidation and reduction reactions, where electrons are transferred from an electron donor molecule to an electron acceptor. An example of such an enzyme is pyruvate dehydrogenase. Common cofactors for oxidoreductase enzymes include NADP+ or NAD+. The general reaction they catalyze is: AH2+B→A+BH2
- Transferases: Transferases are responsible for transferring a specific chemical group from a donor molecule to an acceptor molecule. Transaminases, for instance, transfer an amino group between molecules. Their general reaction is: A–X+B↔B–X+A
- Hydrolases: These enzymes catalyze hydrolysis reactions, breaking bonds using water molecules. An example is pepsin, which hydrolyzes peptide bonds in proteins. The general reaction they oversee is: A–X+H2O→X–OH+A–H
- Lyases: Lyases facilitate reactions that either form or break double bonds without the involvement of hydrolysis or oxidation. An example is aldolase, which operates in glycolysis. Their typical reaction is: A–X+B–Y→A=B+X–Y
- Isomerases: These enzymes catalyze the conversion of a molecule from one isomeric form to another. They assist in intramolecular rearrangements. An example is phosphoglucomutase, which operates in glycogenolysis. The reaction they catalyze is: ACis→A′Trans
- Ligases: Ligases are responsible for joining two large molecules by forming a new chemical bond between them. DNA ligase, for instance, forms a phosphodiester bond between DNA fragments. Their general reaction is: A+B→AB
| Enzyme Class | Examples | Reaction Catalyzed |
|---|---|---|
| 1. Oxidoreductases | Alcohol dehydrogenase (E.C. 1.1.1.1) | Oxidation-Reduction: AH2 + B -> A + BH2 |
| Cytochrome oxidase | ||
| L- and D-amino acid oxidases | ||
| 2. Transferases | Hexokinase (E.C. 2.7.1.1) | Group Transfer: X + B -> A + B – X |
| Transaminases | ||
| Transmethylases | ||
| Phosphorylase | ||
| 3. Hydrolases | Lipase (E.C. 3.1.1.3) | Hydrolysis: A – B + H2O -> AH + BOH |
| Choline esterase | ||
| Acid and alkaline phosphatases | ||
| Pepsin | ||
| Urease | ||
| 4. Lyases | Aldolase (E.C. 4.1.2.7) | Addition-Elimination: A – B + X – Y -> AX – BY |
| Fumarase | ||
| Histidase | ||
| 5. Isomerases | Triose phosphate isomerase (E.C. 5.3.1.1) | Interconversion of Isomers: A -> A’ |
| Retinol isomerase | ||
| Phosphohexose isomerase | ||
| 6. Ligases | Glutamine synthetase (E.C. 6.3.1.2) | Condensation (usually dependent on ATP): A + B -> A – B + ATP -> ADP + Pi |
| Acetyl CoA carboxylase | ||
| Succinate thiokinase |
Besides the primary classification, enzymes can also be categorized based on their location of action:
- Intracellular Enzymes: Also known as endoenzymes, these enzymes operate within the cells where they are produced. They primarily break down large polymers into smaller chains of monomers. Most enzymes in plants and animals fall under this category. Upon cell death, these enzymes undergo intracellular digestion.
- Extracellular Enzymes: Termed as exoenzymes, these enzymes are secreted by living cells and function outside the cell but within its environment. They mainly act as digestive enzymes, breaking down complex macromolecules into simpler forms for absorption by the cell. Examples include enzymes in bacteria, fungi, and certain insectivores. Unlike intracellular enzymes, exoenzymes undergo external digestion during cell death.
Therefore, understanding the classification of enzymes provides a comprehensive insight into their diverse roles and mechanisms in biological systems. Besides, it emphasizes the importance of enzymes in maintaining the metabolic balance and overall functioning of organisms.
How Enzymes Work? – Mechanism of Enzyme Action
Enzymes play a pivotal role in biological systems as powerful catalysts. Their primary function is to facilitate chemical reactions, ensuring they occur efficiently and effectively. The mechanism of enzyme action can be understood through the following detailed and sequential explanation:
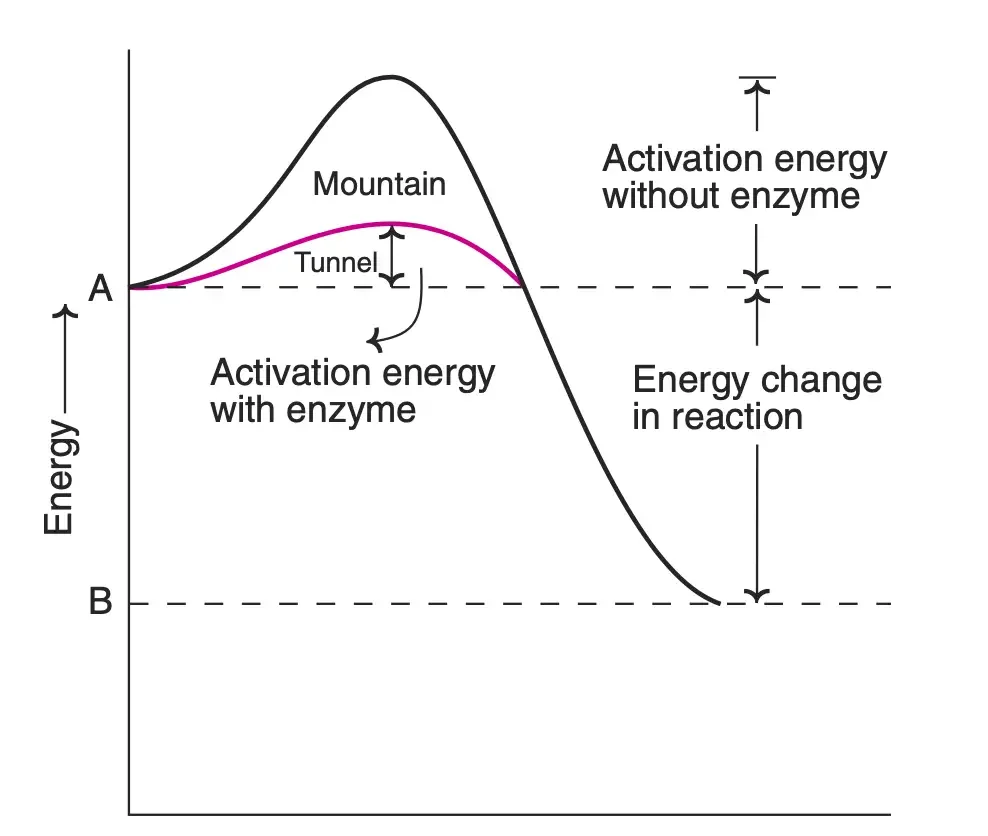
- Activation Energy and Catalysis: For a chemical reaction to transpire, the reactants must reach an activated or transition state. This state requires a certain amount of energy, known as the activation energy. Enzymes function to lower this activation energy, allowing reactions to proceed even at body temperatures below 40°C. Therefore, while enzymes do not alter the equilibrium constants of reactions, they significantly enhance the reaction velocity.
- Enzyme-Substrate Complex Formation: The fundamental requirement for enzyme catalysis is the formation of an enzyme-substrate complex. The substrate (S) binds to the enzyme (E) at its active site, forming this complex (ES). This interaction ultimately leads to the production of the desired product (P). The equation representing this is: E + S → ES → E + P.
- Lock and Key Model: Proposed by Emil Fischer, this model suggests that the enzyme’s structure is rigid. The substrate fits into the enzyme’s binding site, much like a key fits into a lock. However, this model’s rigidity fails to explain the flexible nature of enzymes and the effects of allosteric modulators.
- Induced Fit Theory: Introduced by Koshland in 1958, this theory posits that the enzyme’s active site is not rigid. The substrate’s interaction with the enzyme induces a fit or a conformational change, leading to the formation of a strong substrate binding site. This model, supported by X-ray diffraction studies, also accounts for the action of allosteric modulators and competitive inhibition on enzymes.
- Substrate Strain Theory: This theory suggests that the substrate undergoes strain due to the induced conformational change in the enzyme. The strained substrate then leads to product formation. It is believed that a combination of the induced fit model and substrate strain is operative in enzymatic action.
In conclusion, enzymes work by providing an alternate pathway for reactions, reducing the activation energy required. They bind to substrates, undergo conformational changes, and facilitate the conversion of substrates to products. The detailed mechanisms, including the lock and key model, induced fit theory, and substrate strain theory, provide insights into the intricate processes that govern enzyme action. The use of technical vocabulary, such as “allosteric modulators” and “competitive inhibition,” further emphasizes the complexity and specificity of these biological catalysts.
Enzymes as Biochemical Catalysts
Enzymes, often referred to as biochemical catalysts, play a pivotal role in the field of biochemistry, facilitating a wide array of chemical reactions within living organisms. This phenomenon, known as biochemical catalysis, is fundamental to numerous biological processes. Here, we delve into the world of enzymes as biochemical catalysts, exploring their functions and providing examples of enzyme catalysis.
Functions of Enzymes:
Enzymes are remarkable biological molecules renowned for their ability to accelerate chemical reactions without being consumed or permanently altered during the process. These functions are crucial for life, and they encompass various essential roles:
- Acceleration of Reactions: Enzymes speed up chemical reactions by lowering the activation energy required for the reaction to occur. This enables cellular processes to proceed at biologically relevant rates.
- Specificity: Enzymes exhibit remarkable specificity, meaning they catalyze particular reactions with precision. Each enzyme is designed to work with specific substrates, ensuring that the correct reactions occur in the right cellular contexts.
- Regulation: Enzyme activity can be regulated to maintain cellular homeostasis. Cells can control enzyme activity through feedback mechanisms, allosteric regulation, and other means.
- Metabolism: Enzymes are central to metabolic pathways, including glycolysis, the citric acid cycle, and photosynthesis. These pathways involve a series of enzyme-catalyzed reactions that transform one molecule into another, ultimately producing energy and cellular building blocks.
Examples of Enzyme Catalysis:
- Cane Sugar Inversion: In the food industry, enzymes like invertase are used to convert cane sugar (sucrose) into its constituent sugars, glucose, and fructose. This process, known as cane sugar inversion, is essential for the production of various sweet products.
- C12H22O11(aq) + H2O(l) → C6H12O6(aq) + C6H12O6(aq)
- Conversion of Milk to Curd: The transformation of milk into curd is a classic example of enzyme action. Lactase, an enzyme released by lactobacilli bacteria, catalyzes the conversion of lactose in milk into lactic acid, leading to the curdling of milk.
- Conversion of Glucose into Ethyl Alcohol: Enzymes like zymase play a pivotal role in the fermentation process, converting glucose into ethyl alcohol (ethanol) and carbon dioxide. This process is utilized in the production of alcoholic beverages and biofuel production.
- C6H12O6(aq) → 2C2H5OH(aq) + 2CO2(aq)
- Conversion of Starch into Maltose: Diastase, an enzyme, catalyzes the hydrolysis of starch molecules into maltose, a disaccharide composed of two glucose molecules. This process is significant in the production of malted beverages and food products.
Enzymes are not only vital in industrial applications but also in the intricate biochemical reactions that sustain life. Their specificity, efficiency, and ability to function under mild conditions make enzymes indispensable tools in various fields, from food production to pharmaceuticals and biotechnology. Therefore, the study of enzymes as biochemical catalysts is essential for understanding and harnessing their potential in diverse applications.
Mechanism Of Enzyme Catalysis
Enzyme catalysis is a fundamental process in biological systems, ensuring that biochemical reactions occur at an accelerated rate. The formation of an enzyme-substrate complex (ES) is pivotal for this catalysis to take place and subsequently produce the desired products. Remarkably, reactions catalyzed by enzymes are estimated to be 10^6 to 10^12 times faster than those that are not catalyzed. This significant enhancement in reaction rate can be attributed to several processes:
- Acid-base Catalysis: Acids and bases play a pivotal role in enzymology. At physiological pH, histidine is a key amino acid. Its protonated form acts as an acid, while its conjugate functions as a base. Other acids include the OH group of tyrosine, SH group of cysteine, and ε-amino group of lysine. Their conjugates, along with carboxyl ions (COO–), act as bases. A classic example demonstrating the role of acid and base in catalysis is ribonuclease, which cleaves phosphodiester bonds in RNA.
- Substrate Strain: As previously discussed, inducing strain on the substrate during ES formation is essential. This strain elevates the energy level of the substrate, transitioning it to an activated state. The action mechanism of lysozyme, an enzyme found in tears that cleaves β-1,4 glycosidic bonds, is believed to combine both substrate strain and acid-base catalysis.
- Covalent Catalysis: Here, the enzyme’s active site contains either a negatively charged (nucleophilic) or positively charged (electrophilic) group. This group attacks the substrate, leading to a covalent bond between the substrate and enzyme. Serine proteases, named for the presence of serine at their active site, exemplify enzymes where covalent catalysis and acid-base catalysis occur simultaneously. Examples include chymotrypsin, trypsin, and thrombin.
- Proximity Catalysis: For effective catalysis, reactants must be in close proximity to the enzyme. The higher the substrate concentration, the faster the reaction rate. As enzymes bind to substrate molecules at their active sites, catalysis can increase exponentially, often by at least a thousand-fold.
In conclusion, enzyme catalysis is a multifaceted process, often involving a combination of acid-base catalysis, substrate strain, covalent catalysis, and proximity catalysis. These processes collectively facilitate the substrate’s transition to an activated state, leading to product formation. The use of technical vocabulary, such as “nucleophilic” and “electrophilic,” underscores the intricate nature of these biological catalysts.
Thermodynamics Of Enzymatic Reactions
Thermodynamics plays a pivotal role in understanding the energy dynamics of enzymatic reactions. Based on energy considerations, enzymatic reactions can be categorized into three distinct types:
- Isothermic Reactions: In these reactions, the energy exchange between the reactants and products is minimal, meaning there’s negligible energy difference between the starting materials and the end products. An illustrative example of this type of reaction is the action of glycogen phosphorylase: Glycogen+Pi→Glucose 1-phosphateGlycogen+Pi→Glucose 1-phosphate
Here, the conversion of glycogen to glucose 1-phosphate occurs without any significant energy change. - Exothermic (Exergonic) Reactions: These reactions are characterized by the release of energy. In other words, the products have lower energy than the reactants. A classic example of an exothermic enzymatic reaction is the breakdown of urea by the enzyme urease: Urea→NH3+CO2+energyUrea→NH3+CO2+energy
In this reaction, the decomposition of urea results in the formation of ammonia, carbon dioxide, and the liberation of energy. - Endothermic (Endergonic) Reactions: Contrary to exothermic reactions, endothermic reactions require an input of energy. The products of these reactions possess higher energy than the reactants. An example of this type of reaction is the phosphorylation of glucose by the enzyme glucokinase: Glucose+ATP→Glucose 6-phosphate+ADPGlucose+ATP→Glucose 6-phosphate+ADP
In this scenario, glucose is converted to glucose 6-phosphate, and this transformation necessitates energy, which is sourced from ATP.
In conclusion, the thermodynamics of enzymatic reactions provides a comprehensive understanding of the energy dynamics involved in various biochemical processes. Whether energy is conserved, released, or consumed during these reactions offers insights into the intricate balance and efficiency of biological systems. The use of technical terms, such as “isothermic,” “exothermic,” and “endothermic,” emphasizes the scientific rigor and specificity of these processes.
Enzyme-Substrate Interactions
Enzyme-substrate interactions play a crucial role in the realm of biochemistry, where enzymes, large proteinaceous compounds, act as biocatalysts to facilitate essential reactions within living organisms. These reactions are integral to various life processes, and enzymes aid substrates in navigating their transformation. To understand this intricate dance between enzymes and substrates, one must delve into the molecular details of their interactions.
Enzymes are equipped with specific features on their outer surfaces, such as functional groups like -SH and -COOH. These features create hollow spaces that act as binding sites for substrates. The concept is akin to a key fitting into a lock, as substrates with charges opposite to those on the enzyme’s active site find their place within these spaces. This specific substrate binding site is termed the enzyme’s active site (E).
One favored model explaining enzyme-substrate interaction is known as the induced-fit model. According to this model, the interaction between the substrate and enzyme begins as a weak one. These initial weak interactions initiate rapid conformational changes in both the substrate and enzyme, strengthening their binding. This process also brings catalytic sites into close proximity to the substrate’s bonds, facilitating the subsequent chemical reactions.
Several mechanisms underlie the process of catalysis within enzyme-substrate interactions:
- Catalysis by Bond Strain: This mechanism involves induced structural rearrangements that strain substrate bonds, making it easier for them to reach the transition state. The new conformation forces substrate atoms and catalytic groups, such as aspartate, into configurations that stress substrate bonds.
- Covalent Catalysis: In covalent catalysis, substrates are oriented in a way that a covalent intermediate forms between the enzyme and substrate. For example, serine proteases illustrate this mechanism in proteolysis. The active site serine’s hydroxyl group generates a covalent bond with a peptide bond’s carbonyl carbon, resulting in peptide bond hydrolysis.
- Catalysis Involving Acids and Bases: Some mechanisms complement strain-based catalysis. For instance, the use of glutamate as a general acid catalyst aids in catalytic events.
- Catalysis by Orientation and Proximity: Enzyme-substrate interactions align reactive groups in close proximity, facilitating their involvement in catalysis. Reactive groups like aspartate, when positioned near the substrate, are more likely to participate in catalytic reactions.
Understanding these mechanisms sheds light on the remarkable precision and efficiency of enzymatic reactions, highlighting their central role in the biochemical processes that sustain life.
Action and Nature of Enzymes
Enzymes, the remarkable biological catalysts, play a pivotal role in facilitating a wide array of biochemical reactions with exceptional precision and efficiency. To comprehend the action and nature of enzymes, it is crucial to delve into the intricate details of their mechanism.
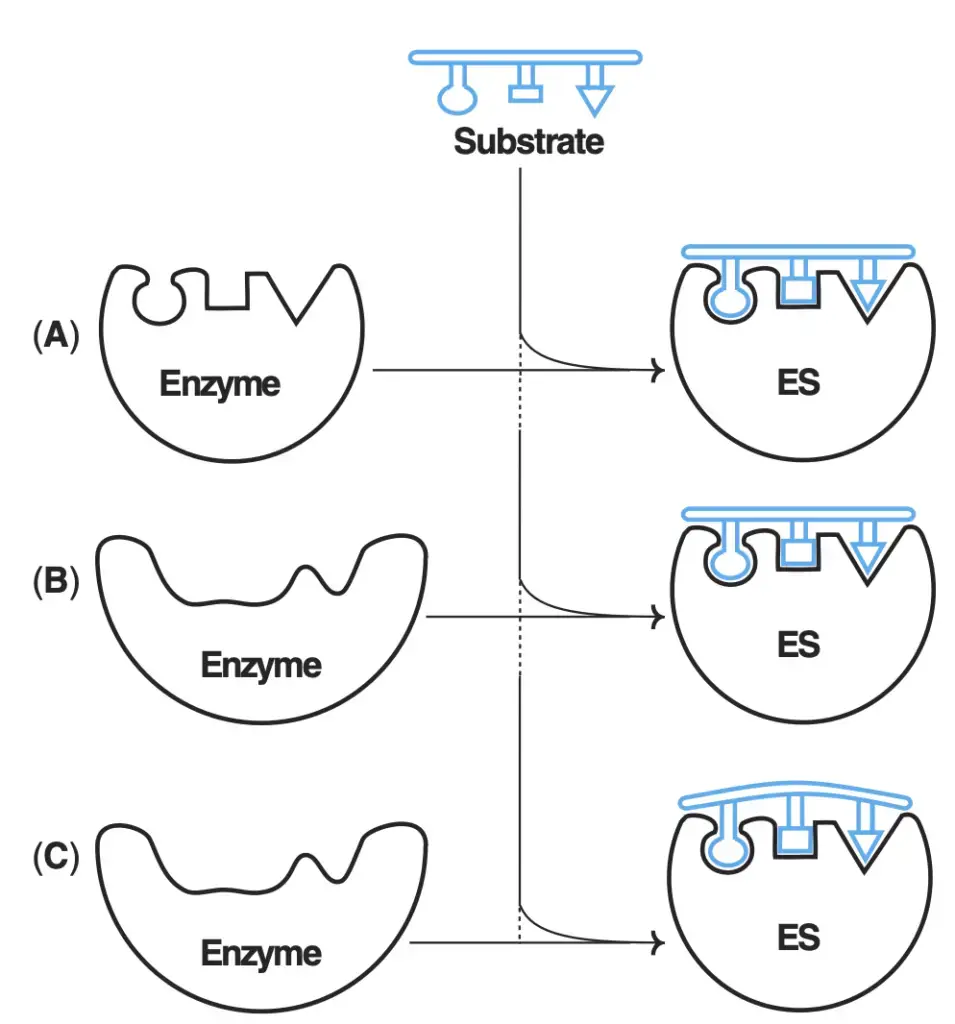
Enzymes operate through a highly specific and structured process. It all begins with the enzyme’s active site, a region designed to accommodate a particular substrate (S) with a matching structure. This specificity is akin to a lock-and-key mechanism, ensuring that only the correct substrate can fit into the active site. As the substrate binds to the enzyme’s active site, an intermediate complex known as ES (enzyme-substrate) is formed.
The critical function of the enzyme lies in its ability to lower the activation energy required for a chemical reaction to occur. By providing a suitable surface for the substrate, enzymes facilitate the transition state of the reaction. This transition state represents a moment of unstable equilibrium when the substrate undergoes chemical transformations, forming new bonds and breaking existing ones.
The enzymatic action unfolds in two distinct but interconnected steps:
Step 1: Combining of Enzyme and Substrate
E + S → [ES]
In this step, the enzyme (E) and the substrate (S) form an enzyme-substrate complex [ES], marking the initiation of the reaction. The binding of the substrate to the enzyme’s active site is a specific and crucial event.
Step 2: Disintegration of the Complex to Yield the Product
[ES] → E + P
In the second step, the enzyme-substrate complex [ES] undergoes disintegration, leading to the formation of the product (P) and the release of the enzyme (E). During this process, the enzyme remains unchanged and available for further reactions, perpetuating the catalytic cycle.
This catalytic cycle continues as free enzymes bind to other substrates, initiating new reactions. Ultimately, the enzyme’s action leads to the conversion of substrates into products, with the enzyme itself remaining unchanged and available for reuse.
In summary, enzymes serve as highly specific biological catalysts that facilitate biochemical reactions by lowering the activation energy barrier. Their action involves the formation of enzyme-substrate complexes, the transition state of reactions, and the subsequent release of products and free enzymes. This process allows enzymes to play a central role in the intricate web of life’s biochemical processes.
Factors Affecting Enzyme Activity
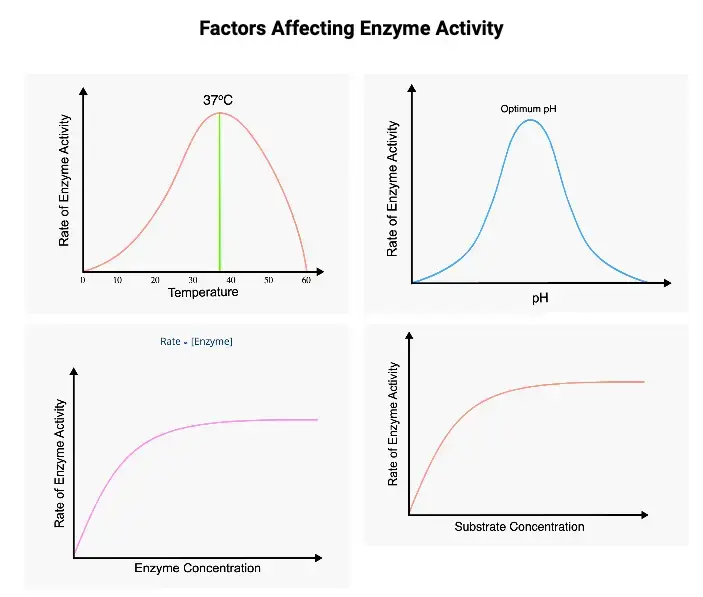
Enzyme activity is a highly regulated process influenced by various factors that play a crucial role in determining the velocity of enzyme reactions. Understanding these factors is essential for comprehending enzymatic function. Here, we explore the key factors affecting enzyme activity:
- Concentration of Enzyme:
- The concentration of the enzyme is a critical factor affecting enzyme activity. Increasing the enzyme concentration proportionately increases the reaction velocity. This property is harnessed in laboratory assays to determine serum enzyme levels for disease diagnosis.
- Concentration of Substrate:
- The concentration of the substrate also significantly impacts enzyme activity. Within a limited range of substrate levels, increasing substrate concentration leads to an increase in reaction velocity. A plot of velocity against substrate concentration results in a rectangular hyperbola, illustrating three distinct phases: linear, curved, and almost unchanged.
- The order of reaction depends on the substrate concentration. When the reaction rate is proportional to the substrate concentration ([S] is less than Km), it’s considered first-order with respect to substrate. If [S] is much greater than Km, the reaction rate becomes independent of substrate concentration, resulting in a zero-order reaction.
- Effect of Temperature:
- Enzyme activity is temperature-dependent. As temperature increases, the velocity of enzyme reactions rises, reaching a maximum before declining. Typically, a bell-shaped curve is observed.
- The temperature coefficient or Q10 represents the increase in enzyme velocity when the temperature is raised by 10°C. For most enzymes, Q10 is around 2 between 0°C and 40°C. Higher temperatures enhance molecular collision and interaction, leading to faster reaction rates.
- Enzymes have an optimal temperature range for activity, usually between 35°C to 40°C. However, some enzymes, like Taq DNA polymerase and muscle adenylate kinase, remain active even at very high temperatures (e.g., 100°C). Most enzymes denature and become inactive above 70°C.
- Effect of pH:
- Changes in hydrogen ion concentration (pH) significantly influence enzyme activity. Each enzyme has an optimum pH at which its activity is maximal. Deviations from this pH result in reduced enzyme activity, with complete inactivity at extreme pH levels.
- Most enzymes in higher organisms exhibit optimal activity around neutral pH (pH 6-8). However, exceptions exist, such as pepsin (active at pH 1-2), acid phosphatase (active at pH 4-5), and alkaline phosphatase (active at pH 10-11). Fungal and plant enzymes are often most active in acidic pH (pH 4-6).
- Hydrogen ions influence enzyme activity by modifying the ionic charges on amino acids, substrates, and enzyme-substrate complexes at the active site.
- Effect of Product Concentration:
- The accumulation of reaction products can inhibit enzyme velocity. For some enzymes, products form a loose complex with the active site, leading to inhibition.
- In living systems, product inhibition is typically prevented by removing products as they are formed.
- Effect of Activators:
- Certain enzymes require inorganic metallic cations (e.g., Mg2+, Mn2+, Zn2+) or anions (e.g., chloride ion) as activators for optimum activity. These ions function as activators through various mechanisms, including substrate binding, ES-metal complex formation, direct participation in reactions, or conformational changes in the enzyme.
- Effect of Time:
- The time required for an enzyme reaction is influenced by various factors, primarily alterations in pH and temperature under ideal conditions.
- Effect of Light and Radiation:
- Exposure to ultraviolet, beta, gamma, and X-rays can inactivate certain enzymes due to the formation of peroxides. For example, UV rays inhibit the activity of salivary amylase.
| Factor | Description |
|---|---|
| Concentration of Enzyme | Increasing enzyme concentration proportionately increases reaction velocity. |
| Concentration of Substrate | Within a limited range, increasing substrate concentration leads to increased reaction velocity. |
| Temperature | Enzyme activity is temperature-dependent, with an optimal temperature range for each enzyme. |
| pH | Enzyme activity is pH-dependent, with each enzyme having an optimum pH for maximal activity. |
| Product Concentration | Accumulation of reaction products can inhibit enzyme velocity. |
| Activators | Some enzymes require specific metallic cations or anions as activators for optimal activity. |
| Time | Enzyme reaction time is influenced by factors such as pH and temperature under ideal conditions. |
| Light and Radiation | Exposure to certain types of light and radiation can inactivate enzymes. |
Regulation Of Enzyme Activity In The Living System
In the intricate web of biological systems, the regulation of enzyme activity stands as a cornerstone for maintaining cellular homeostasis and ensuring efficient metabolic processes. The living system employs a multifaceted approach to regulate enzyme activities, ensuring that cellular functions are carried out with precision and economy. Here’s a detailed exploration of the various mechanisms through which enzyme activities are modulated:
- Allosteric Regulation: This is a pivotal mechanism where enzymes are regulated by molecules that bind to sites other than the active site. These molecules, known as allosteric modulators, can either enhance or inhibit enzyme activity. The binding of these modulators induces conformational changes in the enzyme structure, thereby influencing its activity.
- Activation of Latent Enzymes: Some enzymes are synthesized in an inactive form, known as proenzymes or zymogens. These latent enzymes require specific triggers, such as the cleavage of a peptide bond, to become active. This mechanism ensures that the enzyme is activated only when required, preventing unnecessary metabolic reactions.
- Compartmentation of Metabolic Pathways: To achieve efficiency, cells compartmentalize metabolic pathways, ensuring that related enzymes are localized within specific cellular structures or organelles. This spatial organization allows for a streamlined flow of substrates and products, minimizing unwanted interactions and maximizing metabolic efficiency.
- Control of Enzyme Synthesis: The synthesis of enzymes is intricately regulated at the transcriptional and translational levels. Depending on the cellular needs, the production of specific enzymes can be upregulated or downregulated, ensuring that the right amount of enzyme is present at the right time.
- Enzyme Degradation: Just as the synthesis of enzymes is controlled, their degradation is equally crucial. Cells have specialized mechanisms, like the ubiquitin-proteasome pathway, to degrade and recycle enzymes that are damaged, non-functional, or no longer needed. This ensures that only functional enzymes are present in the cell, contributing to cellular economy.
- Isoenzymes: Isoenzymes, or isozymes, are different forms of the same enzyme that catalyze the same reaction but differ in their amino acid sequences and properties. They provide a layer of regulation, allowing cells to fine-tune enzyme activity based on specific conditions or tissue types.
1. Allosteric regulation and allosteric inhibition
In the realm of enzymology, allosteric regulation plays a pivotal role in modulating enzyme activity. Allosteric enzymes are unique in that they possess additional binding sites, known as allosteric sites, apart from the conventional active sites. These sites are distinct regions on the enzyme molecule where specific substances, termed allosteric effectors, bind and influence enzyme activity.
- Allosteric Effectors: These are specific molecules that bind to the allosteric sites of an enzyme. Their binding can either enhance or inhibit enzyme activity. When a positive (+) allosteric effector, also known as an activator, binds, it increases the enzyme’s activity. Conversely, a negative (-) allosteric effector acts as an inhibitor, binding to the inhibitor site and reducing enzyme activity.
- Classes of Allosteric Enzymes: Based on the influence of the allosteric effector on the enzyme’s Km (substrate affinity) and Vmax (maximum reaction velocity), allosteric enzymes are categorized into two classes:
- K-class: In this class, the effector alters the Km value without affecting the Vmax. The resulting plots resemble those seen in competitive inhibition. An example is phosphofructokinase.
- V-class: Here, the effector impacts the Vmax without changing the Km. The plots obtained are akin to non-competitive inhibition, as seen in enzymes like acetyl CoA carboxylase.
- Conformational Changes: Allosteric enzymes often undergo structural changes upon effector binding. Most allosteric enzymes are oligomeric, meaning they consist of multiple subunits. The binding of an effector molecule induces a conformational shift in the enzyme’s active site, leading to either activation or inhibition. The concerted model posits that allosteric enzymes toggle between two states: the T (tense) and R (relaxed) states. These states exist in a delicate equilibrium, with allosteric inhibitors favoring the T state and activators, along with substrates, leaning towards the R state.
- Feedback Regulation: A hallmark of metabolic pathways is the feedback regulation mechanism. In this process, the final product of a series of enzyme-catalyzed reactions inhibits the first step, ensuring efficient pathway regulation. This type of control, often termed negative feedback regulation, ensures that when end product levels rise, its synthesis decreases, exemplifying cellular economy.
- Feedback Inhibition: A subset of feedback regulation, feedback inhibition involves the direct inhibition of an enzyme by the end product. This mechanism is quintessential for efficient cellular functioning. A classic example is Aspartate transcarbamoylase (ATCase), an enzyme involved in pyrimidine biosynthesis. When the end product, Cytidine triphosphate (CTP), accumulates, it allosterically inhibits ATCase, showcasing a feedback mechanism in action.
In essence, allosteric regulation and inhibition are sophisticated mechanisms that ensure enzymes function optimally, maintaining cellular balance and efficiency. The emphasis on the functions of these regulatory processes, combined with the use of technical vocabulary, underscores the intricate nature of these biological phenomena.
2. Activation of latent enzymes
Latent enzymes are inherently inactive in their original form. However, certain mechanisms activate these enzymes, ensuring they perform their designated functions.
- Proenzymes or Zymogens: Some enzymes are initially synthesized as proenzymes or zymogens. These proenzymes undergo an irreversible covalent activation process, which involves the breakdown of specific peptide bonds. This transformation results in the activation of the enzyme. For instance, chymotrypsinogen, pepsinogen, and plasminogen are proenzymes that, upon activation, are converted into their active forms: chymotrypsin, pepsin, and plasmin, respectively.
- Reversible Covalent Modifications: Certain enzymes can toggle between active and inactive states based on the body’s requirements. This interconversion is facilitated by reversible covalent modifications. Two primary mechanisms are involved:
- Phosphorylation and Dephosphorylation: This process involves the addition or removal of a phosphate group. For example, glycogen phosphorylase, a muscle enzyme responsible for breaking down glycogen to release energy, exists in two forms. The inactive form, phosphorylase b, can be activated through phosphorylation of its serine residues, resulting in the active form, phosphorylase a. Conversely, the dephosphorylation of phosphorylase a produces the inactive form, phosphorylase b. Other enzymes, such as citrate lyase and fructose 2,6-bisphosphatase, also follow this activation mechanism.
- Oxidation and Reduction of Disulfide Bonds: Some enzymes require the presence of sulfhydryl (SH) groups to be active. These enzymes, including succinate dehydrogenase and urease, are stabilized by substances like glutathione.
- Phosphorylation Variability: Interestingly, while many enzymes are activated upon phosphorylation, others become inactive when phosphorylated. For instance, enzymes like glycogen synthase, acetyl CoA carboxylase, and HMG CoA reductase are active in their dephosphorylated state but become inactive upon phosphorylation.
3. Compartmentation
In the intricate cellular environment, compartmentation plays a pivotal role in ensuring metabolic efficiency. This concept revolves around the idea that certain substances in the body, such as fatty acids and glycogen, undergo both synthesis and degradation. Logically, it would be counterproductive for both these processes to occur simultaneously. Therefore, to achieve maximum metabolic economy, the body has strategically placed synthetic (anabolic) and breakdown (catabolic) pathways in different cellular organelles.
For instance, the enzymes responsible for fatty acid synthesis are localized in the cytosol. In contrast, the enzymes facilitating fatty acid oxidation are housed within the mitochondria. This spatial separation prevents the simultaneous occurrence of opposing metabolic pathways, ensuring that resources are utilized efficiently.
To further elucidate this concept, let’s delve into the distribution of certain enzymes and metabolic pathways across various cellular organelles, as presented in below:
Distribution of Enzymes and Metabolic Pathways in Cellular Organelles
- Cytoplasm: This organelle houses enzymes like aminotransferases and peptidases. Metabolic pathways such as glycolysis, hexose monophosphate shunt, fatty acid synthesis, and purine and pyrimidine catabolism also take place here.
- Mitochondria: This powerhouse of the cell is responsible for fatty acid and amino acid oxidation. It also facilitates the Krebs cycle, urea synthesis, and the electron transport chain, culminating in oxidative phosphorylation.
- Nucleus: The primary function of the nucleus is the biosynthesis of DNA and RNA.
- Endoplasmic Reticulum (Microsomes): This organelle plays a role in protein biosynthesis, triacylglycerol and phospholipid synthesis, steroid synthesis and reduction, and houses enzymes like cytochrome P450 and esterase.
- Lysosomes: These are equipped with a plethora of enzymes, including lysozyme, phosphatases, phospholipases, hydrolases, proteases, lipases, and nucleases.
- Golgi Apparatus: This organelle contains enzymes like glucose 6-phosphatase and 5-nucleotidase. It also facilitates the transfer of glucose and galactose molecules.
- Peroxisomes: These organelles house enzymes such as catalase, urate oxidase, and D-amino acid oxidase. They also play a role in the oxidation of long-chain fatty acids.
In conclusion, compartmentation ensures that metabolic pathways operate in specific cellular locations, preventing counterproductive simultaneous reactions and optimizing the use of cellular resources. This strategic distribution underscores the body’s intricate design and its emphasis on metabolic efficiency.
4. Control of enzyme synthesis
In the intricate realm of cellular metabolism, enzymes play a pivotal role in catalyzing and regulating biochemical reactions. The concentration of these enzymes, especially the rate-limiting ones, is typically low. However, it’s crucial to understand that the amount of a specific enzyme directly influences the speed or velocity of the reaction it catalyzes. Interestingly, many rate-limiting enzymes possess short half-lives, which facilitates the efficient regulation of their levels.
Diving deeper into the categorization of enzymes, there are two primary types:
- Constitutive Enzymes (House-keeping Enzymes): These enzymes maintain a relatively constant level, irrespective of external factors. Their concentrations are not modulated and remain stable.
- Adaptive Enzymes: The concentrations of these enzymes are dynamic. They can increase or decrease based on the body’s requirements and are meticulously regulated. The synthesis of these enzymes, which are essentially proteins, is governed by genes, as detailed in Chapter 26.
A crucial aspect of enzyme synthesis control is the phenomena of induction and repression:
- Induction: This term denotes the increased synthesis of an enzyme.
- Repression: In contrast, repression signifies the decreased synthesis of an enzyme.
Both induction and repression play a pivotal role in determining enzyme concentration at the gene level, often mediated by hormones or other specific substances.
Examples of Enzyme Induction: The hormone insulin is a prime example of enzyme induction. It promotes the synthesis of enzymes like glycogen synthetase, glucokinase, phosphofructokinase, and pyruvate kinase. All these enzymes play a vital role in glucose utilization. Another hormone, cortisol, induces the synthesis of several enzymes, including pyruvate carboxylase, tryptophan oxygenase, and tyrosine aminotransferase.
Examples of Repression: There are instances where a substrate can inhibit or repress the synthesis of a particular enzyme. A classic example is pyruvate carboxylase, a key enzyme facilitating the synthesis of glucose from non-carbohydrate sources like pyruvate and amino acids. When there’s an abundance of glucose, the body doesn’t require additional synthesis. This is adeptly managed by the repression of pyruvate carboxylase by glucose itself.
In conclusion, the control of enzyme synthesis is a meticulously orchestrated process, ensuring that the body’s metabolic needs are met efficiently. Through mechanisms like induction and repression, the body can adapt and respond to varying internal and external conditions, emphasizing the dynamic and adaptive nature of cellular metabolism.
5. Enzyme degradation
In the intricate world of cellular biology, enzymes play a pivotal role in facilitating and regulating various biochemical reactions. However, it’s essential to understand that enzymes are not perpetual entities; they undergo degradation over time. This degradation process is vital, as the perpetual existence of enzymes could lead to a plethora of cellular complications.
The longevity or half-life of enzymes exhibits significant variability. For instance:
- LDH4: This enzyme has a half-life ranging between 5 to 6 days.
- LDH1: Its half-life is relatively shorter, spanning 8 to 12 hours.
- Amylase: This enzyme’s half-life is even more transient, lasting between 3 to 5 hours.
It’s noteworthy that key and regulatory enzymes, which play crucial roles in metabolic pathways, are often the most rapidly degraded. Their degradation mechanism is finely tuned to the body’s needs. When these enzymes are not required, they undergo swift degradation, ensuring they don’t accumulate unnecessarily. Conversely, when the body demands these enzymes, they are rapidly synthesized, ensuring timely availability.
An interesting observation is that, in many cases, enzymes with longer half-lives tend to exhibit slower catalytic activities. However, this isn’t a universal truth and may vary based on specific enzymes and their roles.
In conclusion, enzyme degradation is a meticulously regulated process, ensuring that enzymes don’t over-accumulate and potentially disrupt cellular functions. This dynamic balance between synthesis and degradation ensures that enzymes are available when needed and removed when they are not, emphasizing the adaptive and responsive nature of cellular metabolism.
6. Isoenzymes
- In the realm of biochemistry, enzymes play a pivotal role in catalyzing and regulating various biochemical reactions. Among these enzymes, a unique category known as isoenzymes or isozymes emerges. Isoenzymes are essentially multiple forms of the same enzyme, and their existence serves a specific purpose in the regulation of enzyme activity.
- One of the defining characteristics of isoenzymes is their tissue-specific nature. This means that certain isoenzymes predominantly exist or function within specific tissues or organs. Therefore, the presence or abundance of a particular isoenzyme in a tissue can provide insights into the metabolic activities of that tissue.
- While isoenzymes catalyze the same biochemical reaction, they exhibit differences in certain kinetic properties. Specifically, they may differ in their Km (Michaelis constant) and Vmax (maximum reaction velocity). The Km value represents the substrate concentration at which the reaction rate is half of its maximum, while Vmax is the maximum rate achieved by the system. Variations in these values among isoenzymes can influence the efficiency and rate of the reaction they catalyze.
- For instance, lactate dehydrogenase (LDH) and creatine phosphokinase (CPK) are two enzymes that have distinct isoenzyme forms. These isoenzymes, while performing the same fundamental reaction, can have different Km and Vmax values, leading to variations in their catalytic efficiencies.
- In conclusion, isoenzymes are a testament to the intricate and adaptive nature of cellular metabolism. Their existence allows for fine-tuned regulation of enzymatic reactions, ensuring that metabolic processes are optimized for the specific needs of different tissues. Through their varied kinetic properties, isoenzymes ensure that biochemical reactions proceed efficiently and effectively, catering to the diverse metabolic demands of the body.
Units Of Enzyme Activity
Enzymes, the biological catalysts, play a pivotal role in various biochemical reactions. However, when it comes to expressing the quantity of enzymes, it’s not about their concentration, such as in milligrams or micrograms. Instead, the emphasis is on their activity. Over the years, various methods have been developed to estimate enzyme activities, especially for plasma enzymes. As a result, enzyme activities have been expressed in diverse ways, including King-Armstrong units, Somogyi units, Reitman-Frankel units, and spectrophotometric units, to name a few.
- Katal: A Standardized Unit To bring about uniformity in the expression of enzyme activities worldwide, the Enzyme Commission of the International Union of Biochemistry (IUB) introduced significant changes. They introduced a new unit called “katal,” abbreviated as “kat.” One katal represents the conversion of one mole of substrate per second (mol/sec). Furthermore, the activity can be expressed in varying scales such as millikatals (mkat) or microkatals (µkat).
- International Units (IU): The SI Standard Some researchers and professionals opt for the standard units or the System International (SI) units. One International Unit (IU) is defined as the enzyme activity that facilitates the conversion of one micromole of substrate per minute. It’s noteworthy that the SI units and katal are interconvertible. Specifically, 1 IU is equivalent to 16.67 nkat, and conversely, 1kat equals 60 million IU.
- Laboratory Practices and Enzyme Units Despite the introduction of standardized units like katal or SI units, many clinical laboratories continue to employ traditional units such as King-Armstrong units or Somogyi units. Therefore, when expressing enzyme activities, it becomes imperative to specify the units of enzyme activity. This ensures clarity and facilitates comparison, especially when referencing normal values.
In conclusion, while enzymes are integral to biochemical processes, the way their activity is expressed has seen variations over the years. The introduction of standardized units like katal and SI units aims to bring about uniformity. However, the transition to these units in clinical settings is still underway, emphasizing the importance of always specifying the units when discussing enzyme activities.
What is the Active Site of Enzyme?
The active site of an enzyme is a fundamental component responsible for its catalytic activity. Here is an informative description of the active site based on the provided content:
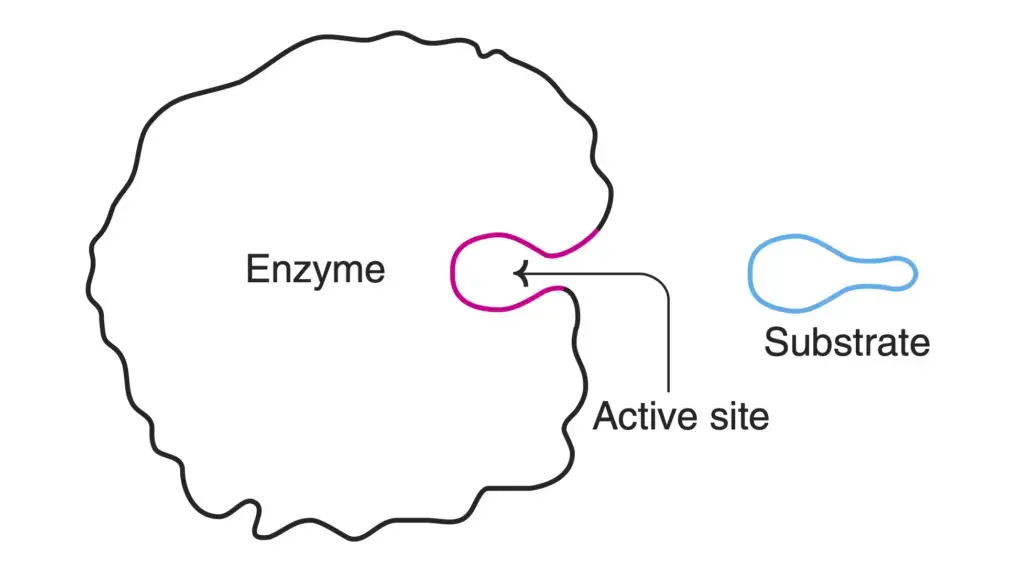
Enzymes, despite their relatively large size compared to substrates, have a specific region within their molecular structure that plays a critical role in binding substrates and catalyzing reactions. This region is known as the active site (or active center) of an enzyme, and it is where the enzyme interacts directly with its substrate(s) to facilitate chemical reactions.
Key characteristics and features of the active site include:
- Tertiary Structure: The active site’s existence is a result of the unique three-dimensional native conformation of the enzyme’s tertiary structure.
- Amino Acid Composition: The active site comprises specific amino acids, often referred to as catalytic residues. These amino acids are not necessarily adjacent in the linear sequence of amino acids that make up the enzyme’s primary structure. For instance, in the enzyme lysozyme, the active site is formed by contributions from amino acid residues with different numerical positions (e.g., 35, 52, 62, 63, and 101).
- Cleft or Pocket: Active sites are often described as clefts, crevices, or pockets within the larger enzyme molecule, occupying a relatively small region.
- Flexibility: The active site is not rigid but rather flexible, allowing it to adapt and accommodate specific substrate molecules.
- Substrate Binding and Catalysis: Typically, the active site consists of both a substrate binding site and a catalytic site. The catalytic site is responsible for facilitating the specific chemical reaction.
- Cofactors and Coenzymes: Some enzymes require cofactors or coenzymes for their activity, and these may also be part of the active site.
- Weak Noncovalent Bonds: Substrates bind to the active site through weak noncovalent bonds, ensuring that the enzyme-substrate complex can form and dissociate as needed.
- Specificity: Enzymes exhibit specificity in their function, primarily due to the presence and characteristics of their active sites.
- Common Amino Acids: Amino acids frequently found at active sites include serine, aspartate, histidine, cysteine, lysine, arginine, glutamate, and tyrosine. Serine, in particular, is commonly encountered.
- Formation of Enzyme-Substrate Complex: Substrates bind to the enzyme at the active site, forming an enzyme-substrate complex (ES). Following catalysis, the product (P) is released, and the enzyme is available for reuse.
Understanding the active site is crucial for comprehending the specificity, functionality, and efficiency of enzyme-catalyzed reactions.
Enzyme Specificity
Enzyme specificity is a fundamental characteristic that sets enzymes apart from chemical catalysts. Enzymes exhibit a remarkable degree of specificity in their actions, allowing them to perform highly selective and precise functions within biological systems. This specificity can be categorized into three distinct types: stereospecificity, reaction specificity, and substrate specificity.
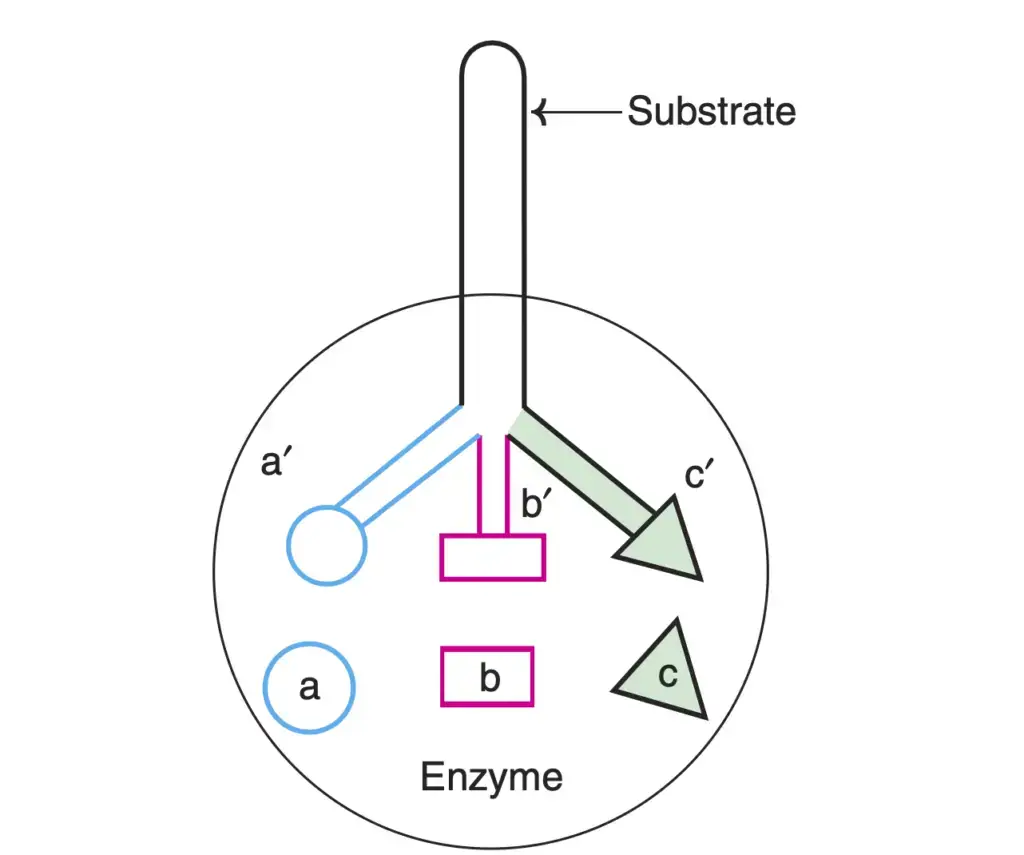
- Stereospecificity (Optical Specificity):
- Stereoisomers are compounds with the same molecular formula but differing structural configurations.
- Enzymes display stereospecificity by acting exclusively on one stereoisomer.
- Examples include:
- L-amino acid oxidase and D-amino acid oxidase, which act on L- and D-amino acids, respectively.
- Hexokinase, which acts on D-hexoses.
- Glucokinase, which acts specifically on D-glucose.
- Amylase, which cleaves α-glycosidic linkages.
- Cellulase, which cleaves β-glycosidic bonds.
- Stereospecificity is attributed to the complementary binding of specific regions of the substrate molecule to corresponding regions on the enzyme’s surface.
- Isomerases, a class of enzymes, do not exhibit stereospecificity as they are involved in interconverting isomers.
- Reaction Specificity:
- Enzymes demonstrate reaction specificity when the same substrate can undergo different types of reactions, each catalyzed by a distinct enzyme.
- For instance, amino acids can undergo various reactions such as transamination, oxidative deamination, decarboxylation, racemization, etc., with each reaction catalyzed by a different enzyme.
- This phenomenon highlights the diversity of enzymatic reactions within the biological system.
- Substrate Specificity:
- Substrate specificity pertains to the selectivity of enzymes for specific substrates.
- It can be categorized into three types:
- Absolute Substrate Specificity: Certain enzymes act exclusively on a single substrate. For example, glucokinase acts solely on glucose, converting it to glucose 6-phosphate, while urease cleaves urea into ammonia and carbon dioxide.
- Relative Substrate Specificity: Some enzymes act on structurally related substances, often dependent on specific functional groups or bonds. For instance, trypsin exhibits group specificity by hydrolyzing peptide linkages involving arginine or lysine, while chymotrypsin cleaves peptide bonds attached to aromatic amino acids like phenylalanine, tyrosine, and tryptophan. Bond specificity is also observed, such as glycosidases acting on glycosidic bonds of carbohydrates or lipases cleaving ester bonds of lipids.
- Broad Substrate Specificity: Certain enzymes can act on closely related substrates, a phenomenon known as broad substrate specificity. An example is hexokinase, which acts on glucose, fructose, mannose, and glucosamine but not on galactose. This suggests that some structural similarity among the first four compounds makes them common substrates for hexokinase.
Enzyme specificity plays a pivotal role in maintaining the precision and efficiency of biochemical reactions in living organisms. It ensures that each enzyme fulfills a specific function within complex metabolic pathways, contributing to the overall functioning of biological systems.
What are Cofactor?
Enzyme cofactors are essential chemical components that play a critical role in the catalytic activity of enzymes. These cofactors are non-protein molecules or ions that associate with enzymes to facilitate and enhance their function. The interaction between enzymes and cofactors is crucial for catalyzing specific biochemical reactions. Here, we will explore the various types of cofactors found in enzymes and their functions:
Types of Cofactors Present in Enzymes:
- Prosthetic Groups: Prosthetic groups are cofactors that are tightly and permanently bound to the enzyme. They are an integral part of the enzyme’s structure and function. These groups play a vital role in catalysis and are involved in specific chemical reactions. For example, many enzymes contain a prosthetic group known as FAD (Flavin Adenine Dinucleotide), which is covalently linked to the enzyme. FAD participates in redox reactions by accepting and donating electrons during catalysis, making it an essential component for these enzyme functions.
- Coenzymes: Coenzymes are non-protein organic molecules that transiently interact with enzymes during catalytic reactions. Unlike prosthetic groups, coenzymes can dissociate from the enzyme after the reaction is complete. Coenzymes serve as carriers of specific chemical groups or participate in the transfer of atoms or functional groups between molecules. One of the most well-known coenzymes is NAD+ (Nicotinamide Adenine Dinucleotide), which plays a crucial role in redox reactions by accepting and donating electrons, thus facilitating energy transfer in various metabolic pathways.
- Metal Ions: Metal ions, such as zinc (Zn2+), magnesium (Mg2+), iron (Fe2+), and copper (Cu2+), act as cofactors in some enzymes. These metal ions are often found in the active sites of enzymes and play essential roles in catalysis. They can stabilize reaction intermediates, participate in electron transfer reactions, and facilitate substrate binding. For instance, Zn2+ ions are involved in the catalytic activity of several enzymes, including metalloenzymes, where they assist in coordinating substrate molecules and promoting specific chemical reactions.
Functions of Cofactors in Enzymes:
Cofactors are indispensable for the proper functioning of enzymes. They contribute to enzyme activity in the following ways:
- Catalysis: Cofactors act as catalysts themselves, aiding in the acceleration of chemical reactions. They provide an active site for substrates to bind and undergo specific chemical transformations. For example, NAD+ acts as a coenzyme in redox reactions, facilitating the transfer of electrons between molecules and enabling the conversion of substrates into products.
- Stabilization: Cofactors, especially metal ions, can stabilize reaction intermediates by coordinating with specific functional groups on substrates. This stabilization lowers the activation energy required for the reaction, making it more favorable.
- Substrate Binding: Cofactors often play a role in substrate binding, enhancing the affinity of enzymes for their respective substrates. This interaction ensures that the enzyme specifically targets its intended substrate.
In summary, cofactors in enzymes are diverse and essential components that contribute to the catalytic efficiency of these biological molecules. They include prosthetic groups, coenzymes, and metal ions, each with distinct roles in facilitating enzymatic reactions. These cofactors enable enzymes to carry out specific biochemical transformations efficiently, contributing to the myriad of metabolic processes essential for life.
What are Coenzymes?
Coenzymes are essential components that collaborate with enzymes to facilitate and enhance their catalytic activity. While enzymes, primarily composed of proteins, play a central role in catalyzing biochemical reactions, they often require non-protein cofactors, referred to as coenzymes, to fulfill their catalytic functions. Coenzymes can be classified as either organic or inorganic in nature and are typically low molecular weight and dialyzable substances.
Key points about coenzymes:
- Holoenzyme and Apoenzyme:
- The functional enzyme, known as the holoenzyme, is composed of two essential components: the protein part called the apoenzyme and the non-protein part, which is the coenzyme.
- When a non-protein moiety is tightly bound to the enzyme and is not easily separable by dialysis, it is referred to as a prosthetic group.
- Inorganic cofactors, such as Ca2+, Mg2+, Mn2+, are referred to as activators and are necessary to enhance enzyme activity.
- Some authors may use the terms cofactor, coenzyme, and prosthetic group interchangeably.
- Coenzymes as Second Substrates:
- Coenzymes are often considered “second substrates” or co-substrates because they exhibit an affinity for the enzyme similar to that of the primary substrates.
- During enzymatic reactions, coenzymes undergo chemical alterations, but they are later regenerated, allowing them to participate in multiple reaction cycles.
- This stands in contrast to the primary substrate, which is ultimately converted into the product.
- Participation in Various Reactions:
- Coenzymes play a crucial role in various enzymatic reactions that involve the transfer of atoms or functional groups such as hydrogen, aldehyde, keto, amino, acyl, methyl, carbon dioxide, and more.
- These coenzymes actively facilitate the transfer of these groups, making them essential for enzyme function.
- Derivatives of B-Complex Vitamins:
- Many coenzymes are derivatives of water-soluble B-complex vitamins.
- The biochemical functions of B-complex vitamins are largely exerted through their respective coenzymes.
- These coenzymes are involved in a wide range of metabolic processes, and their structural details and functions are discussed in the context of vitamin-related coenzymes.
- Non-Vitamin Coenzymes:
- Not all coenzymes are derived from vitamins. Some organic substances function as coenzymes independently of vitamin-related pathways.
- Examples of non-vitamin coenzymes include ATP, CDP, UDP, and others.
- These coenzymes play distinct roles in various enzymatic reactions and are not directly related to vitamin metabolism.
- Nucleotide Coenzymes:
- Certain coenzymes possess a structure resembling nucleotides, with nitrogenous bases, sugars, and phosphate groups.
- Examples of nucleotide coenzymes include NAD+, NADP+, FMN, FAD, coenzyme A, UDPG, among others.
- These coenzymes are involved in critical metabolic processes and serve as carriers of specific functional groups.
- Protein Coenzymes:
- In some cases, proteins can act as coenzymes. One example is thioredoxin, a protein that serves as a coenzyme for the enzyme ribonucleotide reductase.
- These protein coenzymes play essential roles in specific enzymatic reactions.
- Coenzyme Role in Enzyme Specificity:
- While coenzymes are crucial for enzyme function, they do not primarily determine enzyme specificity.
- Enzyme specificity is predominantly determined by the apoenzyme, or the protein part of the enzyme, rather than the coenzyme.
- Coenzymes may participate in multiple enzymatic reactions, and their specificity is largely dictated by the enzyme they interact with.
| Coenzyme (Abbreviation) | Derived from | Atom or Group | Dependent Enzyme |
|---|---|---|---|
| Thiamine pyrophosphate (TPP) | Thiamine | Aldehyde or Keto | Transketolase |
| Flavin mononucleotide (FMN) | Riboflavin | Hydrogen and Electron | L-Amino Acid Oxidase |
| Flavin adenine dinucleotide (FAD) | Riboflavin | Hydrogen and Electron | D-Amino Acid Oxidase |
| Nicotinamide adenine dinucleotide (NAD+) | Niacin | Hydrogen | Lactate Dehydrogenase |
| Nicotinamide adenine dinucleotide phosphate (NADP+) | Niacin | Hydrogen | Glucose 6-Phosphate Dehydrogenase |
| Lipoic acid | Lipoic Acid | Pyruvate Dehydrogenase Complex | |
| Pyridoxal phosphate (PLP) | Pyridoxine | Amino or Keto | Alanine Transaminase |
| Coenzyme A (CoA) | Pantothenic Acid | Acyl | Thiokinase |
| Tetrahydrofolate (FH4) | Folic Acid | One Carbon | Formyl Transferase (Formyl, Methenyl, etc.) |
| Biocytin | Biotin | CO2 | Pyruvate Carboxylase |
| Methylcobalamin; Deoxyadenosyl Cobalamin | Cobalamin | Methyl/Isomerisation | Methylmalonyl CoA Mutase |
In summary, coenzymes are indispensable partners to enzymes, facilitating and regulating a wide array of biochemical reactions within living organisms. They are critical components of enzyme systems, working together to ensure the precise and efficient execution of metabolic processes.
| Coenzyme | Abbreviation | Biochemical Functions |
|---|---|---|
| Adenosine triphosphate | ATP | Donates phosphate, adenosine, and adenosine monophosphate (AMP) moieties. |
| Cytidine diphosphate | CDP | Required in phospholipid synthesis as a carrier of choline and ethanolamine. |
| Uridine diphosphate | UDP | Carrier of monosaccharides (glucose, galactose), required for glycogen synthesis. |
| S-Adenosylmethionine | SAM | Donates methyl group in biosynthetic reactions (active methionine). |
| Phosphoadenosine phosphosulfate | PAPS | Donates sulfate for the synthesis of mucopolysaccharides (active sulfate). |
What are Non-protein Enzymes?
In the vast realm of biological catalysts, enzymes predominantly stand out as protein molecules that accelerate biochemical reactions. However, an intriguing exception to this norm is the existence of non-protein enzymes. Among these, ribozymes are a notable category that warrants attention.
Ribozymes: The RNA Catalysts
Ribozymes are specialized ribonucleic acids (RNAs) that possess the unique ability to function as biological catalysts. Contrary to the common perception of enzymes being proteins, ribozymes challenge this notion by being non-protein in nature. Their discovery and understanding have added a new dimension to the field of enzymology.
In 1983, a groundbreaking revelation was made by Altman and his team. They were studying ribonuclease P, an enzyme previously believed to cleave precursors of tRNAs to produce tRNAs. Their research unveiled that the enzyme’s functionality was attributed to the RNA component present within it, rather than its protein segment. This was a significant departure from the established understanding, as detailed in Chapter 4.
Upon isolating the RNA part from ribonuclease P, it was observed that this RNA segment displayed genuine enzyme activity. Furthermore, it adhered to the Michaelis-Menten kinetics, a foundational concept in enzyme kinetics. Subsequent research fortified the idea that RNA molecules can indeed serve as enzymes and facilitate catalysis.
Structural Adaptability of RNA
Just as proteins adopt specific conformations to function effectively, RNA molecules too exhibit a similar behavior. They can assume a tertiary structure, which is believed to be instrumental in their role as biocatalysts. This specific conformation of RNA might be the key to its enzymatic function.
A Glimpse into Evolutionary Perspectives
Delving into the evolutionary context, it is postulated that ribozymes, or RNA catalysts, played a pivotal role before the emergence of protein enzymes. This suggests that during the early stages of evolution, ribozymes might have been the primary catalysts driving biochemical reactions.
In conclusion, ribozymes stand as a testament to the diverse and intricate world of biological catalysts. Their existence as non-protein enzymes offers valuable insights into the evolutionary history and the multifaceted nature of enzymatic reactions in living organisms.
Applications Of Enzymes
Enzymes, as biological catalysts, play a pivotal role in various biochemical processes. Their significance extends beyond just biological systems, finding applications in therapeutic treatments, analytical procedures, genetic engineering, and industrial processes. This expository piece aims to elucidate the diverse applications of enzymes based on the provided content.
1. Therapeutic Applications of Enzymes: Enzymes have found a prominent place in the realm of medical treatments. Some notable therapeutic applications include:
- Streptokinase and Urokinase: Derived from streptococcus, streptokinase is instrumental in dissolving blood clots. It functions by activating plasma plasminogen, which subsequently transforms fibrin into soluble products, thereby clearing clots.
- Asparaginase: This enzyme plays a crucial role in leukemia treatments. Tumor cells rely on the host’s plasma asparagine for proliferation. Administering asparaginase depletes the plasma’s asparagine levels, thereby inhibiting tumor cell viability.
- Papain: Recognized for its anti-inflammatory properties.
- α1-Antitrypsin: Employed in emphysema treatments.
- Pancreatic Enzymes (trypsin, lipase): These are essential for digestion, especially in pancreatic ailments.
2. Enzymes in Analytical Applications: Enzymes serve as valuable reagents in clinical laboratories, aiding in the precise measurement of substrates, drugs, and even other enzyme activities. For instance:
- Glucose Oxidase and Peroxidase: These enzymes, when immobilized on paper strips, facilitate the detection of glucose in urine. The intensity of the resultant blue hue is indicative of glucose concentration, offering a semi-quantitative assessment.
3. Enzymes in Genetic Engineering: Enzymes have revolutionized genetic engineering practices. Key enzymes in this domain include:
- Restriction Endonucleases: Essential for gene transfer and DNA fingerprinting.
- Taq DNA Polymerase: Central to the polymerase chain reaction.
4. Industrial Applications of Enzymes: The industrial sector harnesses the power of enzymes for various processes:
- Rennin: Employed in cheese production.
- Glucose Isomerase: Used in the synthesis of high fructose syrup.
- α-Amylase and Proteases: Find applications in the food industry and washing powders, respectively.
5. Immobilized Enzymes: For prolonged preservation and enhanced stability, enzymes can be immobilized by binding them to an insoluble matrix. Such immobilized enzymes retain their catalytic activity. Common matrices include beaded gels and cyanogen bromide activated sepharose.
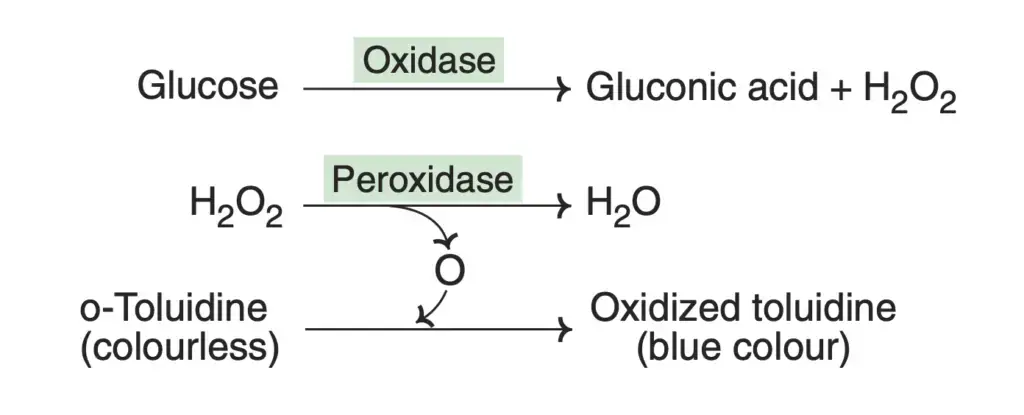
Functions/ Biological roles of Enzymes
Enzymes, as the molecular workhorses of biological systems, serve indispensable functions across a spectrum of vital processes. This expository exploration delves into the diverse biological roles that enzymes play, elucidating their significance based on the provided content.
- Catalysis in Metabolism and Digestion: Enzymes are pivotal in metabolic pathways and digestion. They accelerate chemical reactions that break down complex molecules into simpler forms, facilitating nutrient absorption and energy production.
- Cellular Regulation and Signal Transmission: Enzymes such as kinases and phosphatases are instrumental in cellular regulation and signal transmission. Kinases add phosphate groups to proteins, modulating their activity, while phosphatases remove these groups, thereby regulating cellular responses.
- Metabolic Pathway Regulation: Throughout the body, diverse enzymes orchestrate the regulation of reactions within various metabolic pathways. This meticulous control ensures that intermediate metabolites are synthesized or broken down in response to cellular requirements.
- Negative Feedback Mechanisms: Enzymes play a vital role in negative feedback mechanisms. They activate or inhibit specific reactions to maintain homeostasis. This dynamic adjustment ensures that intermediate metabolites are synthesized or degraded in accordance with cellular demands.
- Post-Translational Modifications: Enzymes catalyze post-translational modifications of proteins. These include phosphorylation, glycosylation, and cleavage of polypeptide chains. These modifications impact protein function, localization, and stability, influencing diverse cellular processes.
- Regulation of Enzyme Levels: Certain enzymes are involved in the regulation of enzyme levels. They control the rate of enzyme degradation, influencing the enzyme’s presence and activity within cells.
- Implications for Health and Disease: Enzymes’ tight regulation is paramount for maintaining homeostasis. Alterations in enzyme structure or production can lead to diseases, disrupting metabolic processes and cellular functions.
- Industrial Applications: Enzymes synthesized by various organisms find practical utility in various industries. They contribute to wine production, cheese production, bread whitening, and fabric design, showcasing their versatility in non-biological domains.
Diagnostic Importance Of Enzymes
Diagnostic medicine harnesses the invaluable role of enzymes, particularly in assessing and prognosticating various diseases. This exposition unveils the diagnostic importance of enzymes, offering a comprehensive understanding of their clinical relevance based on the provided content.
- Plasma Functional Enzymes: Enzymes in circulation can be categorized into two groups: plasma-specific or plasma functional enzymes. These enzymes, often synthesized in the liver, have specific functions and exhibit higher activity levels in plasma than in tissues. Examples include lipoprotein lipase, plasmin, thrombin, choline esterase, and ceruloplasmin.
- Plasma Non-Functional Enzymes: Plasma non-functional enzymes, on the other hand, are either absent or present at low concentrations in plasma compared to their tissue levels. Gastrointestinal digestive enzymes found in plasma, such as amylase, pepsin, trypsin, and lipase, are referred to as secretory enzymes. Constitutive enzymes, such as lactate dehydrogenase, transaminases, and acid and alkaline phosphatases, are associated with cellular metabolism. These enzymes play a crucial role in the diagnosis and prognosis of various diseases.
- Markers of Cellular Damage: Normal serum enzyme levels reflect the equilibrium between their synthesis and release during routine cellular turnover. Elevated enzyme levels can signify cellular damage, increased cell turnover, cell proliferation, or enhanced enzyme synthesis. Serum enzymes serve as biomarkers, aiding in the detection of cellular damage and contributing to disease diagnosis.
- Selected Diagnostic Enzymes: Several enzymes hold diagnostic importance, with their levels serving as indicators of specific diseases. Notable examples include:
- Amylase: Elevated levels indicate acute pancreatitis.
- Alanine Transaminase (ALT/SGPT): Increased levels are associated with acute hepatitis, jaundice, and cirrhosis.
- Aspartate Transaminase (AST/SGOT): Elevated activity suggests myocardial infarction and liver diseases.
- Alkaline Phosphatase (ALP): Increased ALP levels are observed in bone and liver diseases.
- Acid Phosphatase (ACP): Elevated in prostate gland cancer, particularly tartarate-labile ACP.
- Lactate Dehydrogenase (LDH): Used for diagnosing myocardial infarction, infective hepatitis, leukemia, and muscular dystrophy.
- Creatine Kinase (CK): Elevated in early myocardial infarction and muscular dystrophy.
- Decreased Plasma Enzyme Activities: In some disorders, plasma enzyme activities may be lower than normal due to decreased enzyme synthesis or congenital deficiency. Examples include decreased amylase, pseudocholinesterase (ChE II), ceruloplasmin, and glucose 6-phosphate dehydrogenase (G6PD) in RBC.
| Enzymes | Reference value | Disease(s) in which increased | |
|---|---|---|---|
| II. Transaminases | Alanine transaminase (ALT) or serum glutamate pyruvate transaminase (SGPT) | 3–40 IU/l or 40–250 nKat | Acute hepatitis (viral or toxic), jaundice, cirrhosis of liver. |
| Aspartate transaminase (AST) or serum glutamate oxaloacetate transaminase (SGOT) | 4–45 IU/l or 50–320 nKat | Myocardial infarction, liver diseases, liver cancer, cirrhosis of liver. | |
| III. Phosphatases | Alkaline phosphatase (ALP) (pH optimum 9–10) | In adults-3–13 King Armstrong (KA) units/dl In children–15–30 KA/dl | Bone diseases (related to higher osteoblastic activity)-rickets, Pagets’ disease, hyperparathyroidism, carcinoma of bone. Liver diseases-obstructive jaundice (cholestasis), infective hepatitis, cirrhosis of liver. |
| Acid phosphatase (ACP) (pH optimum 4–6) | 0.5–4 KA units/dl | Prostatic carcinoma (tartarate labile ACP serves as a marker for diagnosis and follow-up), Pagets’ disease. | |
| IV. Enzymes of carbohydrate metabolism | Aldolase | 2–6 IU/l | Muscular dystrophy, liver diseases, myocardial infarction, myasthenia gravis, leukemias |
| Isocitrate dehydrogenase (ICD) | 1–4 IU/l | Liver diseases (inflammatory toxic or malignant) | |
| Lactate dehydrogenase (LDH) | 50–200 IU/l or 1–5 Kat | Myocardial infarction, acute infective hepatitis, muscular dystrophy, leukemia, pernicious anemia. | |
| V. Miscellaneous enzymes | Creatine kinase (CK) or creatine phosphokinase (CPK) | 10–50 IU/l | Myocardial infarction (CK useful for early detection), muscular dystrophy, hypothyroidism, alcoholism. |
| Cholinesterase (ChEI) | 2–10 IU/l | Nephrotic syndrome, myocardial infarction. | |
| 5-Nucleotidase or nucleotide phosphatase (NTP) | 2–15 IU/l | Hepatitis, obstructive jaundice, tumors. | |
| Gamma-Glutamyl transpeptidase (GGT) | 5–40 IU/l | Alcoholism, infective hepatitis, obstructive jaundice. | |
| Ceruloplasmin (ferrooxidase) | 20–50 mg/dl | Bacterial infections, collagen diseases, cirrhosis, pregnancy. | |
| I. Digestive enzymes | Amylase | 80–180 Somogyi units/dl or 2.5–5.5 Kat | Acute pancreatitis, mumps (acute parotitis), obstruction in pancreatic duct, severe diabetic ketoacidosis. |
| Lipase | 0.2–1.5 IU/l | Acute pancreatitis, moderate elevation in carcinoma of pancreas. |
Isoenzymes
The concept of isoenzymes or isozymes unveils a fascinating realm of enzyme diversity, wherein multiple forms of an enzyme catalyze the same reaction while exhibiting distinctions in various aspects. This expository text meticulously elucidates the intricate world of isoenzymes, adhering to the provided content’s style and objectives.
- Isoenzyme Distinctions: Isoenzymes, also known as isozymes, represent diverse variants of an enzyme that share the capability to catalyze the same chemical reaction. However, they diverge significantly in terms of their physical and chemical properties. These disparities encompass structural differences, electrophoretic and immunological characteristics, Km (Michaelis constant) and Vmax (maximum velocity) values, optimal pH conditions, susceptibility to inhibitors, and denaturation tendencies.
- Explaining Isoenzyme Existence: Several factors contribute to the existence of isoenzymes within living systems:
- Gene Variation: Isoenzymes can be synthesized from different genes, exemplified by the distinct malate dehydrogenase found in the cytosol compared to that in mitochondria.
- Oligomeric Complexity: Oligomeric enzymes composed of more than one type of subunit, such as lactate dehydrogenase and creatine phosphokinase, may give rise to isoenzymes.
- Monomeric or Oligomeric Activity: Some enzymes can function as either monomers or oligomers. For instance, glutamate dehydrogenase demonstrates this duality.
- Glycoprotein Variability: Differences in carbohydrate content within glycoprotein enzymes can lead to the formation of isoenzymes, a notable example being alkaline phosphatase.
- Lactate Dehydrogenase (LDH) Isoenzymes: LDH serves as an exemplary subject for the exploration of isoenzymes. LDH, with its systematic name L-lactate-NAD+ oxidoreductase (E.C. 1.1.1.27), facilitates the conversion of lactate to pyruvate. Five distinct LDH isoenzymes, LDH1 through LDH5, exist. These isoenzymes can be separated through electrophoresis, with LDH1 exhibiting the highest electrophoretic mobility due to its more positive charge. LDH is a tetrameric enzyme comprising four polypeptide subunits, including M (muscle) and H (heart) subunits produced by different genes. The presence of these subunits gives rise to LDH1 through LDH5, each with unique properties outlined in Table.
- Diagnostic Significance of LDH: LDH isoenzymes play a pivotal role in diagnosing heart and liver-related disorders. LDH1 predominates in heart muscle, and its inhibition by pyruvate ensures the conversion of pyruvate to acetyl CoA instead of lactate, suited for aerobic conditions in the heart. In contrast, LDH5 is prevalent in skeletal muscle, with minimal inhibition by pyruvate, favoring lactate production under anaerobic conditions. Monitoring LDH activity is invaluable for clinical diagnosis.
- Creatine Phosphokinase (CK) Isoenzymes: CK, or creatine phosphokinase, participates in the conversion of phosphocreatine to creatine. CK isoenzymes encompass CPK1 (BB), CPK2 (MB), and CPK3 (MM), with CPK2 (MB) demonstrating diagnostic significance in myocardial infarction, where its levels dramatically increase within hours after the event.
- Alkaline Phosphatase (ALP) Isoenzymes: ALP boasts as many as six isoenzymes primarily distinguished by differences in carbohydrate content. Notable ALP isoenzymes include α1-ALP, α2-heat labile ALP, α2-heat stable ALP, pre-β ALP, and γ-ALP. These isoenzymes serve as indicators of various health conditions, including hepatitis and bone diseases.
- Isoenzymes of alcohol dehydrogenase: Alcohol dehydrogenase (ADH) has two heterodimer isoenzymes. Among the white Americans and Europeans, 1 isoenzyme is predominant whereas in Japanese and Chinese (Orientals) 2 is mostly present. The isomer 2 more rapidly converts alcohol to acetaldehyde. Accumulation of acetaldehyde is associated with tachycardia (increase in heart rate) and facial flushing among Orientals which is not commonly seen in whites. It is believed that Japanese and Chinese have increased sensitivity to alcohol due to the presence of 2–isoenzyme of ADH.
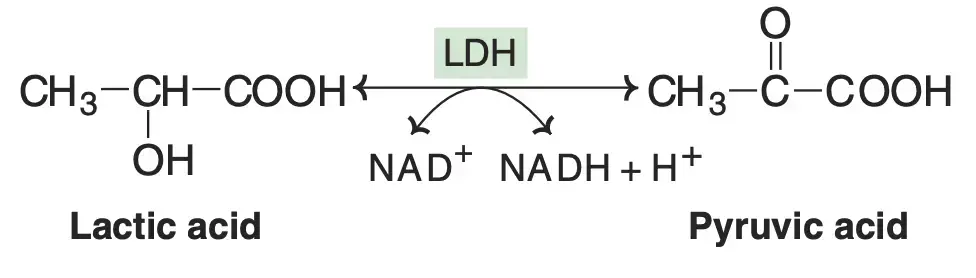
| Isoenzyme | Subunit | Principal Tissue of Origin | Electrophoretic Mobility | Heat Stability (at 60°C) | Percentage of Normal Serum in Humans |
|---|---|---|---|---|---|
| LDH1 | H4 | Heart and RBC | Slowest | No | 25% |
| LDH2 | H3M | Heart and RBC | Slow | No | 35% |
| LDH3 | H2M2 | Brain and Kidney | Fast | Partially | 27% |
| LDH4 | HM3 | Liver and Skeletal Muscle | Faster | Yes | 8% |
| LDH5 | M4 | Skeletal Muscle and Liver | Fastest | Yes | 5% |
In summary, isoenzymes epitomize the intricacies of enzymatic diversity, and their study reveals an array of biochemical adaptations within living systems. Understanding these variations is instrumental in both clinical diagnosis and unraveling the complexities of biological processes.
Enzyme Pattern In Diseases
Enzyme patterns in diseases provide valuable diagnostic information, and it is often more reliable to assess multiple serum enzymes rather than relying on a single enzyme. Here are examples of enzyme patterns in significant diseases:
1. Enzymes in Myocardial Infarction:
- Creatine Phosphokinase (CPK), specifically the MB isoenzyme, is the first enzyme to be released into circulation within 6-18 hours after a myocardial infarction (MI). It is highly useful for early MI diagnosis, reaching its peak value within 24-30 hours and returning to normal levels by the 2nd or 3rd day.
- Aspartate Transaminase (AST or SGOT) rises sharply after CPK and reaches its peak within 48 hours of the MI. AST takes 4-5 days to return to normal levels.
- Lactate Dehydrogenase (LDH1) generally starts rising from the second day after an infarction, reaches its peak by the 3rd or 4th day, and takes about 10-15 days to return to normal levels. LDH is the last enzyme to rise and fall in MI.
- Cardiac Troponins (CT), although not enzymes, are highly useful for early MI diagnosis. Troponin I and Troponin T are essential proteins in this context. Troponin I is released into circulation within four hours after the onset of MI, reaching its peak value by 12–24 hours and remaining elevated for about a week.
- Myoglobin is also an early marker for MI diagnosis but is not specific to cardiac diseases.
- High serum concentration of Brain Natriuretic Peptide is a marker for congestive cardiac failure.
| Diagnostic Marker | Time of Peak Elevation | Time of Return to Normal | Diagnostic Importance |
|---|---|---|---|
| Myoglobin | 4-6 hrs | 20–25 hrs | Earliest marker, however not cardiac-specific. |
| Cardiac Troponin I | 12-24 hrs | 5-9 days | Early marker and cardiac-specific. |
| Cardiac Troponin T | 18-36 hrs | 5-14 days | Relatively early marker and cardiac-specific. However, elevated in other degenerative diseases. |
| Creatine Phosphokinase (MB) | 20-30 hrs | 24-48 hrs | Cardiac-specific and early marker. |
| Lactate Dehydrogenase (LDH I) | 48-72 hrs | 10-15 days | Relatively late marker and cardiac-specific. |
| Aspartate Transaminase | 30-48 hrs | 4-6 days | Not cardiac-specific. |
2. Enzymes in Liver Diseases:
- Elevations in serum levels of Alanine Transaminase, Aspartate Transaminase, and Lactate Dehydrogenase are useful for diagnosing liver dysfunction due to viral hepatitis (jaundice), toxic hepatitis, cirrhosis, and hepatic necrosis.
- In cases of intrahepatic and extrahepatic cholestasis, Alkaline Phosphatase and 5’-Nucleotidase markedly increase.
- Serum-γ-glutamyl Transpeptidase is a valuable marker for alcoholic liver diseases.
3. Enzymes in Muscle Diseases:
- In muscular dystrophies, serum levels of specific muscle enzymes such as Creatine Phosphokinase, Aldolase, and Aspartate Transaminase are increased.
4. Enzymes in Cancers:
- Elevated serum levels of Acid Phosphatase (Tartarate Labile) are specific for detecting prostatic carcinoma. Prostate-specific antigen (PSA) is a more reliable marker for prostate cancer detection.
- Neuron-specific Enolase serves as a marker for lung cancer, neuroblastoma, pheochromocytoma, and other related conditions.
These enzyme patterns play a crucial role in diagnosing and monitoring various diseases, providing valuable insights into the underlying health conditions. Therefore, assessing multiple enzymes allows for a more comprehensive and accurate diagnosis of specific diseases.
| Disease/Enzyme(s) | Significance |
|---|---|
| I Myocardial Infarction | Refer to First Table |
| II Hepatic Disease |
|
| III Muscle Disease |
|
| IV Bone Disease |
|
| V Pancreatic Disease |
|
| VI Prostate Cancer |
|
Diagnostic Importance Of Enzymes In Other Body Fluids And Tissues
Enzymes play a crucial role in diagnosing various medical conditions, and their diagnostic importance extends beyond serum/plasma enzymes to include enzyme estimations in other body fluids and tissues. This broader spectrum of enzyme analysis provides valuable insights into a range of diseases and disorders. Here are some examples of diagnostic importance:
- Urine:
- Urinary amylase levels can be increased in cases of acute pancreatitis, providing a diagnostic clue to this condition.
- The presence of β-N-Acetylgalactosidase in urine at elevated levels may indicate renal graft dysfunction.
- Elevated β-Glucuronidase levels in urine can be associated with cancers of the urinary bladder, pancreas, and other related malignancies.
- Cerebrospinal Fluid (CSF):
- Lactate dehydrogenase levels in cerebrospinal fluid (CSF) can be increased in cases of meningitis, aiding in the diagnosis of this inflammatory condition affecting the brain and spinal cord.
- Gastric Juice:
- β-Glucuronidase activity in gastric juice can be elevated in gastric carcinoma, potentially serving as a diagnostic marker for this type of cancer.
- Feces:
- Fetal trypsin levels in feces can be decreased in individuals with cystic fibrosis, offering diagnostic information about this genetic disorder that affects the respiratory, digestive, and reproductive systems.
- Liver:
- Glucose 6-phosphatase activity in the liver can be significantly lower in individuals with type I glycogen storage disease. This enzyme abnormality helps in diagnosing this rare metabolic disorder characterized by glycogen accumulation.
- Muscle:
- Phosphorylase activity in muscle tissue can be decreased in individuals with McArdle’s disease, a glycogen storage disorder primarily affecting muscle function. This enzyme deficiency aids in the diagnosis of the condition.
- Erythrocytes (Red Blood Cells):
- Glucose 6-phosphate dehydrogenase (G6PD) deficiency in erythrocytes can lead to hemolytic anemia, and this condition is diagnosed by assessing enzyme levels.
- Decreased transketolase activity in erythrocytes can be indicative of thiamine deficiency, a condition that affects energy metabolism and neurological function.
- Cultured Fibroblasts and Amniotic Cells:
- Cultured fibroblasts and amniotic cells are valuable tools for diagnosing inborn errors of metabolism. For instance, phenylalanine hydroxylase deficiency, which leads to phenylketonuria (PKU), can be identified in cultured amniotic cells.
Examples of Enzymes
Enzymes are essential biological molecules that play a crucial role in catalyzing specific chemical reactions within living organisms. These enzymes exhibit remarkable specificity, meaning they are highly selective in the reactions they catalyze. Here are some examples of enzymes and their functions:
- Lipases: Lipases are a group of enzymes that play a vital role in the digestion of lipids (fats) in the intestine. They break down triglycerides, the primary dietary fat, into glycerol and fatty acids. This process is essential for the absorption of dietary fats and their utilization for energy storage. Lipases are produced by the pancreas and secreted into the small intestine.
- Amylase: Amylase is an enzyme responsible for the conversion of complex carbohydrates, such as starch and glycogen, into simpler sugars like glucose and maltose. It is found in various tissues, including saliva, where it begins the digestion of starch in the mouth. Salivary amylase initiates the breakdown of carbohydrates into more digestible forms.
- Maltase: Maltase is an enzyme that specifically targets maltose, a disaccharide composed of two glucose molecules. It catalyzes the hydrolysis of maltose into two glucose molecules. Maltose is present in various foods, including potatoes, pasta, and beer. Maltase helps in the digestion and absorption of maltose-derived glucose in the small intestine.
- Trypsin: Trypsin is a protease enzyme that plays a pivotal role in the digestion of dietary proteins. It breaks down proteins into smaller peptides and amino acids, making them absorbable in the small intestine. Trypsin is produced and released by the pancreas into the small intestine, where it complements the action of other proteases.
- Lactase: Lactase is an enzyme located in the small intestine that is responsible for the digestion of lactose, a sugar found in milk and dairy products. Lactase hydrolyzes lactose into its constituent sugars, glucose and galactose. Individuals with lactose intolerance often have reduced or deficient lactase activity, leading to digestive discomfort when consuming dairy products.
- Helicase: Helicase is an enzyme involved in DNA replication and repair. Its primary function is to unwind or “unzip” the double-stranded DNA molecule by breaking the hydrogen bonds between complementary base pairs. This unwinding process allows other enzymes, like DNA polymerase, to access the DNA strands and perform replication or repair activities.
- DNA Polymerase: DNA polymerase is an essential enzyme responsible for synthesizing new DNA strands during DNA replication. It uses deoxyribonucleotides as building blocks to create a complementary DNA strand based on the template strand. DNA polymerase ensures accurate and faithful replication of the genetic material.
These examples illustrate the diverse functions of enzymes in biological systems. Enzymes are critical for breaking down complex molecules into simpler forms that can be absorbed, utilized, or replicated by living organisms. Their specificity ensures that the right reactions occur at the right time, contributing to the overall functioning and survival of living organisms.
Industrial applications of Enzymes
Enzymes, with their remarkable catalytic properties, find a multitude of applications in various industries, revolutionizing processes and enhancing efficiency. These industrial applications leverage enzymes’ ability to accelerate chemical reactions under mild conditions. Let’s explore some key industrial applications of enzymes:
1. Biofuel Industry:
- Enzymes Used: Cellulases, Ligninases
- Uses: Enzymes like cellulases play a pivotal role in breaking down cellulose into sugars, which can then be fermented to produce cellulosic ethanol. Ligninases are essential for pretreating biomass, preparing it for biofuel production.
2. Biological Detergent:
- Enzymes Used: Proteases, Amylases, Lipases, Mannanases
- Uses: Enzymes in biological detergents, such as proteases, amylases, and lipases, effectively remove protein, starch, and fat or oil stains from laundry and dishware. Mannanases are employed to eliminate food stains from guar gum, a common food additive.
3. Brewing Industry:
- Enzymes Used: Amylase, Glucanases, Proteases, Betaglucanases, Amyloglucosidase, Pullulanases, Acetolactate Decarboxylase (ALDC)
- Uses: Enzymes play a critical role in the brewing industry. They split polysaccharides and proteins in malt, improve wort and beer filtration characteristics, make low-calorie beer, and enhance fermentation efficiency by reducing diacetyl formation.
4. Culinary Uses:
- Enzymes Used: Papain
- Uses: Papain is employed to tenderize meat for cooking, improving its texture and taste.
5. Dairy Industry:
- Enzymes Used: Rennin, Lipases
- Uses: Rennin is used to hydrolyze proteins in the manufacture of cheese, while lipases are crucial in producing specific cheeses like Camembert and Roquefort.
6. Food Processing:
- Enzymes Used: Amylases, Proteases, Trypsin, Cellulases, Pectinases
- Uses: Enzymes play a vital role in food processing, converting starch into sugars (e.g., high-fructose corn syrup production), lowering the protein content of flour (used in biscuit-making), manufacturing hypoallergenic baby foods, and clarifying fruit juices.
7. Molecular Biology:
- Enzymes Used: Nucleases, DNA Ligase, Polymerases
- Uses: In molecular biology, enzymes like nucleases, DNA ligase, and polymerases are instrumental in techniques such as restriction digestion and the polymerase chain reaction (PCR), facilitating the creation of recombinant DNA.
8. Paper Industry:
- Enzymes Used: Xylanases, Hemicellulases, Lignin Peroxidases
- Uses: Enzymes are employed in the paper industry to remove lignin from kraft pulp, a crucial step in paper production.
9. Personal Care:
- Enzymes Used: Proteases
- Uses: Proteases are utilized in personal care products to remove proteins from contact lenses, thereby preventing infections.
10. Starch Industry:
- Enzymes Used: Amylases
- Uses: Amylases are pivotal in converting starch into glucose and various syrups, finding applications in the starch industry.
These examples underscore the versatility of enzymes in various industrial processes, showcasing their indispensable role in optimizing efficiency, enhancing product quality, and minimizing environmental impact. Enzymes continue to drive innovation across a spectrum of industries, making them invaluable tools in modern industrial processes.
| Application | Enzymes Used | Uses |
|---|---|---|
| Biofuel Industry | Cellulases, Ligninases | Break down cellulose into sugars for ethanol production; biomass pretreatment. |
| Biological Detergent | Proteases, Amylases, Lipases, Mannanases | Remove protein, starch, fat, and food stains from laundry and dishware. |
| Brewing Industry | Amylase, Glucanases, Proteases, Betaglucanases, Amyloglucosidase, Pullulanases, Acetolactate Decarboxylase (ALDC) | Split polysaccharides and proteins, improve beer filtration, make low-calorie beer, increase fermentation efficiency. |
| Culinary Uses | Papain | Tenderize meat for cooking. |
| Dairy Industry | Rennin, Lipases | Hydrolyze proteins for cheese production; aid in cheese types like Camembert and Roquefort. |
| Food Processing | Amylases, Proteases, Trypsin, Cellulases, Pectinases | Convert starch into sugars, reduce flour protein, make hypoallergenic baby food, clarify fruit juices. |
| Molecular Biology | Nucleases, DNA Ligase, Polymerases | Facilitate molecular techniques like PCR and recombinant DNA creation. |
| Paper Industry | Xylanases, Hemicellulases, Lignin Peroxidases | Remove lignin from kraft pulp in paper production. |
| Personal Care | Proteases | Remove proteins from contact lenses to prevent infections. |
| Starch Industry | Amylases | Convert starch into glucose and syrups. |
References
- Lewis T, Stone WL. Biochemistry, Proteins Enzymes. [Updated 2023 Apr 24]. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2023 Jan-. Available from: https://www.ncbi.nlm.nih.gov/books/NBK554481/
- https://byjus.com/biology/enzymes/
- https://www.geeksforgeeks.org/enzymes-definition-structure-classification-examples/
- https://rgu-website.s3.ap-south-1.amazonaws.com/wp-content/uploads/2021/12/14130814/20_Sayed-Modinur-Rahaman_Microbiology.pdf
- https://www.basu.org.in/wp-content/uploads/2020/06/Enzymes.pdf