What is Emulsion PCR?
- Emulsion PCR (polymerase chain reaction) is a technique used to amplify multiple DNA templates simultaneously in a single reaction. This method differs from traditional PCR in several key aspects, making it a powerful tool in various scientific and diagnostic applications.
- In traditional PCR, a single template DNA is amplified in a solution containing all the necessary reagents for the reaction. Emulsion PCR, on the other hand, involves the creation of water-in-oil emulsion droplets, with each droplet acting as a separate microreactor for amplification. This allows for the isolation and amplification of individual DNA templates within each droplet.
- The process of emulsion PCR begins with the preparation of emulsion droplets, typically using a microfluidic device. Each droplet contains a single template DNA fragment, along with primers and other reagents necessary for amplification. These droplets are then subjected to thermal cycling, which facilitates the amplification of the DNA templates within them.
- One of the key advantages of emulsion PCR is its ability to amplify picoliter (pL) volumes of DNA, significantly reducing the amount of starting material required. This makes it particularly useful for applications where only limited amounts of DNA are available, such as in forensic analysis or when working with rare samples.
- Another important feature of emulsion PCR is its high throughput nature. By generating thousands or even millions of emulsion droplets, each containing a unique DNA template, researchers can amplify and analyze a large number of DNA sequences in parallel. This makes it an ideal tool for applications such as high-throughput sequencing and single-cell analysis.
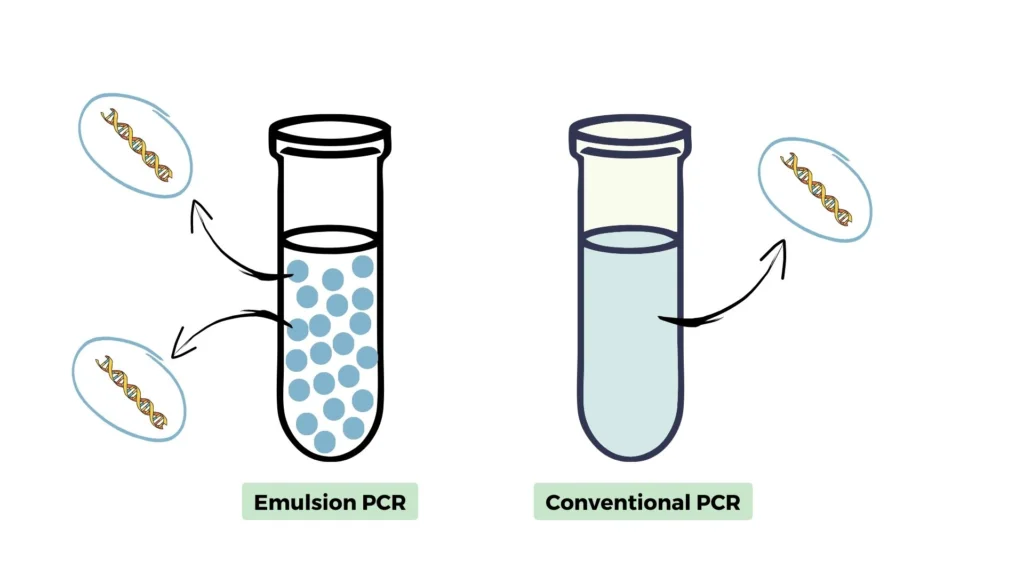
Principle of Emulsion PCR
Emulsion PCR (polymerase chain reaction) is a sophisticated technique designed to amplify individual DNA templates in a high-throughput manner by partitioning the DNA into tiny water-in-oil droplets. Each droplet serves as an independent microreactor, allowing for the separate amplification of distinct DNA fragments. This method leverages the principle of creating isolated environments for DNA amplification, thereby increasing the efficiency and specificity of the process.
The principle of emulsion PCR can be broken down into several key steps:
- DNA Fragmentation: The initial step involves breaking down the DNA into smaller, manageable fragments. This is crucial because it allows for the amplification of specific regions of interest within the DNA sequence. Enzymatic or mechanical methods are typically used for this fragmentation process.
- Emulsion Formation: The fragmented DNA, along with the necessary reagents for PCR (such as primers, nucleotides, and DNA polymerase), is mixed with an oil phase to form a water-in-oil emulsion. This results in the creation of numerous droplets, each containing a single DNA fragment and the reagents needed for amplification. The emulsion acts as a barrier, preventing the mixing of different DNA templates and ensuring that each droplet functions as an isolated reaction chamber.
- Independent Amplification: Within each droplet, the PCR process takes place independently. Thermal cycling is applied to the emulsion, facilitating the denaturation of DNA, annealing of primers, and extension of the new DNA strands. This cycle repeats multiple times, leading to the exponential amplification of the DNA fragment within each droplet.
- Breaking the Emulsion: After the amplification process is complete, the emulsion needs to be broken to recover the amplified DNA. This is typically achieved by adding a demulsifying agent or using centrifugation techniques to separate the oil phase from the aqueous phase.
- Isolation of Amplified DNA Fragments: Once the emulsion is broken, the aqueous phase containing the amplified DNA is collected. At this stage, the DNA is still in a mixture with other components of the reaction, including primers, enzymes, and unused nucleotides.
- Purification: The final step involves purifying the amplified DNA fragments. This can be done using various purification techniques such as column-based purification, magnetic bead separation, or gel electrophoresis. The purified DNA is then ready for downstream applications, such as sequencing, cloning, or further analysis.
The emulsion PCR technique offers several advantages over traditional PCR. By partitioning the DNA into individual droplets, it minimizes cross-contamination and enhances the specificity of the amplification. Additionally, the high-throughput nature of this method allows for the simultaneous amplification of thousands or even millions of DNA fragments, making it an invaluable tool for applications such as high-throughput sequencing and single-cell genomics.
Steps of Emulsion PCR
Emulsion PCR (polymerase chain reaction) is a powerful technique for amplifying DNA by isolating individual DNA fragments within tiny water-in-oil droplets. This compartmentalization ensures that each fragment is amplified independently, increasing efficiency and reducing cross-contamination. The following steps outline the process of emulsion PCR in detail.
1. DNA Fragmentation
- Purpose: To break down the template or genomic DNA into smaller fragments.
- Method: Use restriction enzymes to cut the DNA at specific sequences.
- Ligation: Attach short, known DNA sequences called adapters to the fragments. These adapters serve as primer binding sites for the subsequent PCR steps.
2. Emulsion Preparation
- Components: Mix the aqueous phase (containing the DNA fragments, primers, nucleotides, and DNA polymerase) with the oil phase (mineral or silicon oil) and a surfactant (such as Tween 20).
- Process: Combine these components to create water-in-oil emulsion droplets. Each droplet functions as an independent microreactor, containing all the necessary ingredients for the PCR reaction.
3. PCR Amplification
- Thermal Cycling: Subject the emulsion droplets to thermal cycling. This process involves repeated cycles of:
- Denaturation: Heating to separate the DNA strands.
- Annealing: Cooling to allow primers to bind to the DNA fragments.
- Extension: Heating again to allow DNA polymerase to synthesize new DNA strands.
- Independence: Each droplet contains a single DNA fragment and performs the PCR reaction independently, reducing the chances of cross-contamination and ensuring efficient amplification.
4. Breaking the Emulsion
- Purpose: To release the amplified DNA fragments from the emulsion droplets.
- Methods:
- Centrifugation: Spin the emulsion at high speed to separate the oil and aqueous phases. This method is quick and effective but can potentially disrupt the DNA structure.
- Freezing and Thawing: Freeze and then thaw the emulsion to destabilize and break the droplets.
- Organic Solvent Extraction: Use solvents like isopropanol or ethanol to disrupt the emulsion by destabilizing the water-oil interface. This method is highly effective and yields a high quantity of DNA.
- Detergents: Certain detergents can interact with the surfactants, breaking the emulsion and releasing the DNA.
- Phase Transition Temperature (PTT): Increase the temperature to destabilize the emulsion structure. This method can be effective but carries a risk of DNA denaturation if the temperature is too high.
5. Purification
- Purpose: To remove residual oil, surfactants, enzymes, primers, and other reaction components.
- Techniques:
- Alcohol Precipitation: Use alcohol to precipitate the DNA, then collect it by centrifugation.
- Column-Based Purification: Use columns that selectively bind DNA, allowing contaminants to be washed away.
- Magnetic Bead-Based Purification: Use magnetic beads coated with DNA-binding molecules to capture the DNA, which can then be separated magnetically.
The purified DNA fragments are now ready for downstream applications such as sequencing or cloning. This step-by-step procedure ensures the efficient and accurate amplification of DNA fragments, making emulsion PCR a valuable tool in various scientific and diagnostic applications.
Procedure for the Use of emulsion PCR in NGS
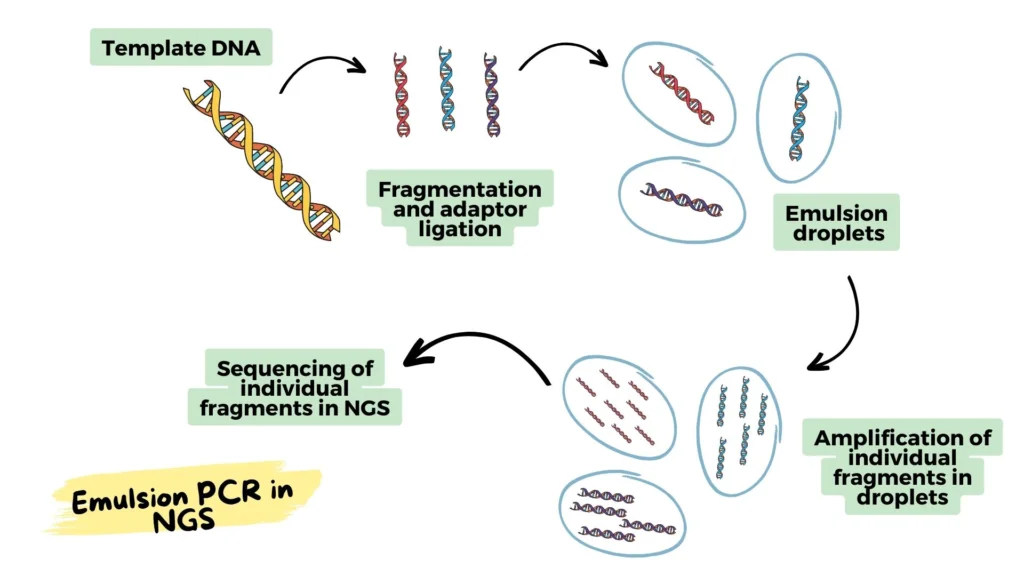
Emulsion PCR is a crucial technique in Next-Generation Sequencing (NGS) due to its ability to amplify large numbers of DNA fragments simultaneously. This step-by-step guide will explain how to incorporate emulsion PCR into the NGS workflow, ensuring accurate and efficient amplification of DNA fragments for sequencing.
Step 1: DNA Isolation
- Method: Use spin-column or automated DNA extraction methods to isolate high-quality DNA from the sample.
- Quality Control: Measure the purity and quantity of the isolated DNA using spectrophotometry or fluorometry to ensure it meets the requirements for downstream applications.
Step 2: DNA Fragmentation
- Process: Fragment the isolated DNA into smaller pieces using restriction enzymes, which cut the DNA at specific sequences.
- Purpose: This step creates manageable-sized fragments suitable for amplification and sequencing.
Step 3: Adaptor Ligation
- Adaptors: Ligate known sequence adaptors to the fragmented DNA. These adaptors are short DNA sequences that provide binding sites for PCR primers.
- Storage: Store the adaptor-ligated DNA fragments in a genomic DNA library for further use.
Step 4: Prepare PCR Mixture
- Ingredients: Add all necessary PCR components (dNTPs, Taq DNA polymerase, primers, and buffer) to the aqueous phase.
- Purpose: Ensure that each droplet in the emulsion contains all the necessary ingredients for DNA amplification.
Step 5: Emulsion Preparation
- Techniques: Create the emulsion by mixing the aqueous phase with the oil phase (mineral or silicon oil) and a surfactant (such as Tween 20). This mixture forms water-in-oil droplets.
- Microreactors: Each droplet acts as an independent microreactor for the PCR reaction.
Step 6: PCR Amplification
- Protocol: Subject the emulsion to a standard PCR protocol, which involves cycles of denaturation, annealing, and extension.
- Independence: Each droplet amplifies a single DNA fragment independently, minimizing cross-contamination and enhancing amplification efficiency.
Step 7: Emulsion Breaking
- Methods: Use centrifugation, freezing and thawing, organic solvent extraction, or detergents to break the emulsion and release the amplified DNA fragments.
- Separation: This step separates the oil phase from the aqueous phase, allowing for the collection of the amplified DNA.
Step 8: DNA Purification
- Techniques: Purify the amplified DNA fragments using alcohol precipitation, column-based purification, or magnetic bead-based purification methods.
- Purpose: Remove any remaining oil, surfactants, enzymes, and other contaminants to obtain clean DNA for sequencing.
Step 9: Sequencing Preparation
- Reaction Setup: Prepare the purified DNA fragments for sequencing by setting up the sequencing reaction according to the specific requirements of the NGS platform being used.
- Sequencing: Load the prepared samples onto the NGS platform and perform the sequencing run.
Step 10: Data Analysis
- Analysis Tools: Use bioinformatics tools to analyze the sequencing data generated.
- Interpretation: Interpret the data to obtain meaningful insights into the DNA sequences, including identifying genetic variations, mutations, and other genomic features.
Advantages of Emulsion PCR
1. Reduction of PCR Duplications
- Individual Amplification: Each DNA fragment is isolated within a water-in-oil droplet, ensuring that amplification occurs independently for each fragment.
- Minimizes Duplications: This isolation reduces the risk of duplicating the same DNA fragment multiple times, leading to more accurate and reliable results.
2. High Throughput
- Massive Parallel Amplification: Emulsion PCR allows for the simultaneous amplification of thousands to millions of DNA fragments within a single reaction.
- Library Enrichment: This high-throughput capability makes ePCR ideal for enriching NGS libraries, significantly increasing the efficiency of sequencing preparations.
3. Sensitivity and Accuracy
- Detection of Low-Abundance Templates: ePCR can amplify DNA fragments present in very low quantities within a sample, making it highly sensitive.
- Accurate Amplification: The compartmentalization of each reaction in droplets reduces the likelihood of contamination and ensures precise amplification of the target sequences.
4. Cost-Effectiveness
- Economical: Emulsion PCR is a cost-effective method for DNA amplification, requiring fewer reagents and resources compared to some other techniques.
- Time-Efficient: The process is relatively quick, allowing for the rapid preparation of DNA libraries for sequencing.
5. Versatility and Precision
- Adaptable: Emulsion PCR can be used with various sequencing platforms, including amplicon sequencing, targeted sequencing, and whole genome sequencing.
- Ease of Use: The technique is straightforward and user-friendly, making it accessible for a wide range of laboratory applications.
- High Precision: By isolating individual DNA fragments in microreactors, ePCR ensures a high level of precision in amplification, which is crucial for accurate downstream analysis.
Limitations of Emulsion PCR
1. Emulsion Preparation Challenges
- Complex Process: Preparing, maintaining, and stabilizing the emulsion throughout the reaction can be complex and challenging.
- Uniform Droplet Size: Ensuring the creation of sufficient and uniform-sized emulsion droplets can be difficult, affecting the consistency of the reaction.
2. Surfactant Interference
- PCR Interference: Surfactants used in emulsion preparation can interfere with PCR amplification, leading to incomplete or partial amplification of DNA fragments.
3. PCR Bias
- Amplicon Size Variability: Emulsion PCR can result in bias, leading to the production of amplicons of varied sizes within the emulsion droplets. This variability can impact the accuracy of the results.
4. Droplet Merging
- Misleading Results: Merging of individual emulsion droplets can occur, leading to the erroneous amplification of DNA fragments and potentially misleading results.
5. Pre- and Post-Processing Steps
- Additional Processing: Emulsion PCR requires additional pre- and post-processing steps, such as emulsion preparation, amplicon separation, and purification, adding complexity and time to the overall process.
Uses of emulsion PCR
1. Next-Generation Sequencing (NGS)
- Library Enrichment: Emulsion PCR is extensively used for library enrichment in NGS workflows.
- High-Quality Libraries: It produces high-quality and high-yield NGS libraries, improving the accuracy and reliability of sequencing results.
2. Single-Cell Genomics
- Cell Heterogeneity Studies: Emulsion PCR is vital for single-cell genomic studies, enabling the amplification of individual fragments of a single cell’s entire genomic DNA.
- Cellular Differences: It helps in understanding the differences and heterogeneity between individual cells.
3. Detection of Rare Variants
- Selective Amplification: Emulsion PCR is valuable in the selective detection of rare variants, which are low-abundance template DNA.
- Effective Amplification: It ensures that rare template DNA, present in very low quantities, is effectively amplified and not missed during conventional PCR.
4. Forensic Analysis
- Trace DNA Detection: Emulsion PCR is used in forensic analysis for the detection of trace amounts of DNA obtained from crime scenes.
- Enhanced Sensitivity: It offers enhanced sensitivity, allowing for the detection of minute amounts of DNA that may be crucial in forensic investigations.
5. Droplet Digital PCR (ddPCR)
- Quantification and Detection: Emulsion PCR is used in ddPCR for more accurate quantification of DNA.
- Mutation Detection: It helps in the detection of mutations and copy number variations with high sensitivity.
- Environmental Studies: Emulsion PCR is also employed in environmental studies for DNA analysis in complex samples.
References
- Kanagal-Shamanna R. Emulsion PCR: Techniques and Applications. Methods Mol Biol. 2016;1392:33-42. doi: 10.1007/978-1-4939-3360-0_4. PMID: 26843044.
- Shao K, Ding W, Wang F, Li H, Ma D, Wang H. Emulsion PCR: a high efficient way of PCR amplification of random DNA libraries in aptamer selection. PLoS One. 2011;6(9):e24910. doi: 10.1371/journal.pone.0024910. Epub 2011 Sep 15. PMID: 21949784; PMCID: PMC3174225.
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.