What is DNA polymerase I?
- DNA polymerase I (Pol I) is an enzyme integral to prokaryotic DNA replication. Identified by Arthur Kornberg in 1956, Pol I was the inaugural DNA polymerase to be discovered, marking a significant milestone in the understanding of polymerases. This enzyme was initially described in the bacterium Escherichia coli (E. coli) and is a common feature in prokaryotic organisms. In many bacteria, including E. coli, the gene encoding for Pol I is termed polA.
- Structurally, the Pol I enzyme in E. coli comprises 928 amino acids. Notably, it exhibits processivity, meaning it can execute multiple consecutive polymerization reactions without detaching from the single-stranded DNA template. While Pol I’s primary role is to assist in the repair of damaged DNA, it also plays a pivotal role in the synthesis of Okazaki fragments. This involves the removal of RNA primers and their subsequent replacement with DNA nucleotides.
- The discovery of Pol I was a result of Arthur Kornberg and his team’s innovative approach, wherein they utilized E. coli extracts to establish a DNA synthesis assay. By incorporating 14C-labeled thymidine, they ensured the production of a radioactive DNA polymer. The purification process involved the addition of streptomycin sulfate, segregating the extract into a nucleic acid-free supernatant (S-fraction) and a nucleic acid-rich precipitate (P-fraction). The latter contained Pol I and other essential factors for DNA synthesis, identified as nucleoside triphosphates. The S-fraction was found to house several deoxynucleoside kinases.
- In recognition of their groundbreaking work on the mechanisms underpinning the biological synthesis of Ribonucleic acid (RNA) and Deoxyribonucleic Acid (DNA), Arthur Kornberg and Severo Ochoa were honored with the Nobel Prize in Physiology or Medicine in 1959.
Definition of DNA polymerase I
DNA polymerase I (Pol I) is an enzyme found in prokaryotes that plays a role in DNA replication and repair, primarily assisting in the removal of RNA primers and filling in the resulting gaps with DNA nucleotides.
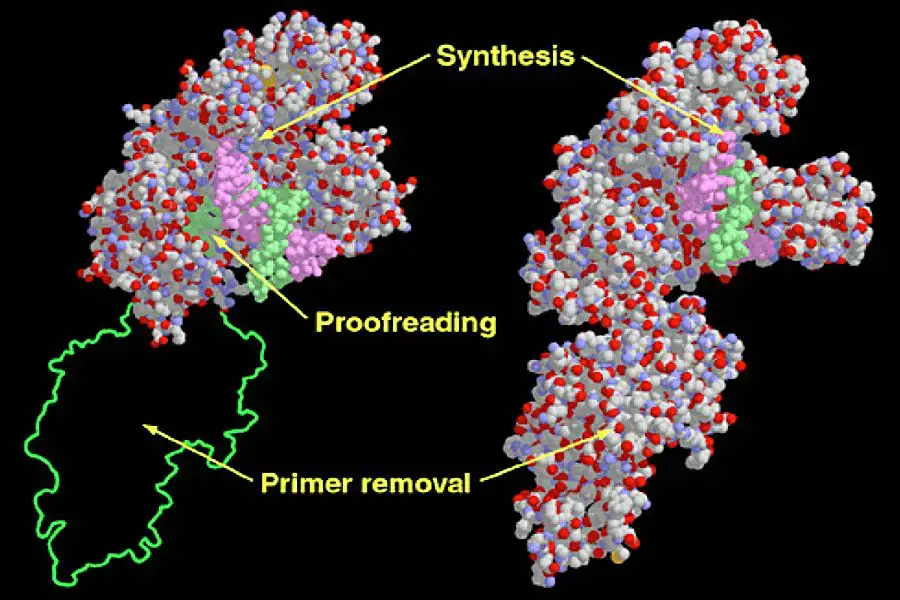
Structure of DNA polymerase I
- DNA polymerase I (Pol I) is an essential enzyme in prokaryotes, primarily involved in DNA repair processes. Structurally, Pol I is categorized under the alpha/beta protein superfamily, characterized by alternating sequences of α-helices and β-strands. The enzyme’s structure in E. coli is intricate, comprising multiple domains that confer three distinct enzymatic activities. These domains, often analogized to a hand’s thumb, finger, and palm, collaborate to facilitate DNA polymerase activity.
- Adjacent to the palm domain, a fourth domain exists, housing an exonuclease active site. This site is responsible for the 3′ to 5′ proofreading mechanism, where incorrectly incorporated nucleotides are excised. Another domain, the fifth, possesses a separate exonuclease active site that operates in a 5′ to 3′ direction, crucial for removing RNA primers during DNA replication or excising DNA during repair processes.
- While E. coli produces five distinct DNA polymerases (Pol I to Pol V), eukaryotic cells are equipped with five different DNA polymerases labeled α, β, γ, δ, and ε. Among these, the eukaryotic DNA polymerase β mirrors E. coli’s DNA Pol I in function, predominantly participating in DNA repair mechanisms like base-excision and nucleotide-excision repairs. To date, 15 distinct human DNA polymerases have been identified.
- For DNA replication, the leading strand is synthesized continuously towards the replication fork, while the lagging strand is synthesized discontinuously in the opposite direction as Okazaki fragments. DNA polymerases necessitate both a template and a primer strand for function. They rely on short RNA segments, or primers, to initiate DNA synthesis. The synthesis then proceeds with the addition of a dNTP to the 3′ hydroxyl group of the existing DNA strand or RNA primer. All DNA polymerases operate through a two-metal ion-catalyzed mechanism, where one metal ion activates the primer’s 3′ hydroxyl group and the other stabilizes the leaving oxygen’s negative charge.
- The X-ray crystal structures of DNA polymerases are often described using a hand analogy. All DNA polymerases encompass three primary domains: the “fingers domain” interacts with the dNTP and template base, the “palm domain” catalyzes the phosphoryl group transfer, and the “thumb domain” engages with the double-stranded DNA. The exonuclease domain, with its unique catalytic site, rectifies mispaired bases. Among the seven DNA polymerase families, the “palm domain” remains conserved in five, while the “finger” and “thumb” domains exhibit variations due to differing secondary structures.
Mechanism of DNA polymerase I
DNA polymerase I (Pol I) plays a specialized role in DNA replication and repair. Its mechanism of action is intricate and ensures the fidelity of DNA synthesis.
- Role in Replication: During DNA replication, especially on the lagging strand, RNA primers are synthesized by primase to initiate the synthesis of Okazaki fragments. Once an Okazaki fragment is synthesized, the RNA primer needs to be removed and replaced with DNA. This is where Pol I comes into play. RNase H removes the RNA primer, and Pol I fills in the resulting gap with the appropriate nucleotides in a 5’→3′ direction.
- Template-Dependent Synthesis: Pol I operates in a template-dependent manner. It can only incorporate nucleotides that form correct base pairs with the template DNA strand. This ensures that the synthesized DNA strand is complementary to the template strand.
- Active Discrimination: Pol I is adept at discriminating between different deoxyribonucleotide triphosphates (dNTPs). Although multiple dNTPs can bind to its active site, Pol I undergoes a conformational change upon binding. This change allows the enzyme to check the geometry and alignment of the base pair formed between the bound dNTP and the template strand. Only the correct A=T and G≡C base pairs fit appropriately in the active site. This mechanism ensures high fidelity during DNA synthesis. However, it’s worth noting that Pol I occasionally makes mistakes, incorporating an incorrect nucleotide once in every 10^4 to 10^5 nucleotides.
- Proofreading Activity: Pol I’s ability to proofread further enhances its fidelity. If an incorrect nucleotide is incorporated, Pol I can recognize the error and correct it, ensuring the accuracy of DNA synthesis.
- Replicative Limitations: Despite its essential functions, Pol I is not the primary enzyme responsible for genome replication in E. coli. Its synthesis rate is between 10 and 20 nucleotides per second, which is considerably slower than the overall replication rate in E. coli. Additionally, Pol I is not highly processive; it tends to dissociate from the DNA after incorporating only 25-50 nucleotides. The discovery of DNA polymerase III, which is more processive and faster, clarified the primary enzyme responsible for genome replication.
In summary, DNA polymerase I is crucial for specific tasks in DNA replication and repair, particularly in filling gaps left by RNA primers and ensuring the accuracy of DNA synthesis through its template-dependent synthesis and proofreading activities.
Functions of DNA polymerase I
DNA polymerase I (Pol I) is a multifunctional enzyme with a suite of enzymatic activities essential for DNA processing. These activities are:
- 5’→3′ DNA-dependent DNA polymerase activity: This is the primary function where Pol I synthesizes DNA in the 5′ to 3′ direction. It requires a primer with a free 3′ hydroxyl group and a DNA template strand to guide the synthesis.
- 3’→5′ exonuclease activity: This reverse activity is responsible for proofreading. As Pol I synthesizes DNA, it checks the newly incorporated nucleotides for accuracy. If a mismatch is detected, this exonuclease activity excises the incorrect nucleotide, ensuring the fidelity of DNA synthesis.
- 5’→3′ exonuclease activity: This forward activity is involved in nick translation, a process crucial during DNA repair. It helps in replacing damaged sections of DNA with the correct sequence.
- 5’→3′ RNA-dependent DNA polymerase activity: While Pol I predominantly operates on DNA templates, it can also function on RNA templates, albeit with significantly reduced efficiency. This activity is believed to have limited biological relevance due to its low efficiency on RNA templates.
Experimental evidence has provided insights into the primary role of Pol I. In a study involving a Pol I-deficient mutant strain of E. coli, it was observed that the strain could still replicate DNA and form colonies, indicating that Pol I is not essential for DNA replication. However, this mutant strain exhibited heightened sensitivity to DNA-damaging agents like UV light. This observation underscored the primary role of Pol I in DNA repair rather than replication. In essence, while Pol I is not pivotal for DNA replication, it plays a crucial role in maintaining genomic integrity by repairing damaged DNA.
Quiz
Who discovered DNA polymerase I?
a) James Watson
b) Francis Crick
c) Arthur Kornberg
d) Rosalind Franklin
[expand title=”Show answer” swaptitle=”Hide answer”] c) Arthur Kornberg [/expand]
Which enzymatic activity does DNA polymerase I NOT possess?
a) 5’→3′ DNA-dependent DNA polymerase activity
b) 3’→5′ exonuclease activity
c) 5’→3′ RNA-dependent RNA polymerase activity
d) 5’→3′ exonuclease activity
[expand title=”Show answer” swaptitle=”Hide answer”] c) 5’→3′ RNA-dependent RNA polymerase activity [/expand]
What is the primary role of DNA polymerase I in DNA replication?
a) Synthesis of the leading strand
b) Removal of RNA primers and filling in the gaps
c) Initiation of DNA replication
d) Joining Okazaki fragments
[expand title=”Show answer” swaptitle=”Hide answer”] b) Removal of RNA primers and filling in the gaps [/expand]
Which domain of DNA polymerase I is responsible for proofreading?
a) Fingers domain
b) Palm domain
c) Thumb domain
d) Exonuclease domain
[expand title=”Show answer” swaptitle=”Hide answer”] d) Exonuclease domain [/expand]
DNA polymerase I operates in which direction for DNA synthesis?
a) 3’→5′
b) 5’→3′
c) Both 3’→5′ and 5’→3′
d) Neither 3’→5′ nor 5’→3′
[expand title=”Show answer” swaptitle=”Hide answer”] b) 5’→3′ [/expand]
In which organism was DNA polymerase I first characterized?
a) Human
b) Yeast
c) E. coli
d) Mouse
[expand title=”Show answer” swaptitle=”Hide answer”] c) E. coli [/expand]
Which of the following is NOT a function of DNA polymerase I?
a) DNA repair
b) DNA replication
c) Transcription
d) Removal of RNA primers
[expand title=”Show answer” swaptitle=”Hide answer”] c) Transcription [/expand]
DNA polymerase I has a synthesis rate of approximately how many nucleotides per second?
a) 1-5
b) 10-20
c) 50-100
d) 500-1000
[expand title=”Show answer” swaptitle=”Hide answer”] b) 10-20 [/expand]
Which domain of DNA polymerase I interacts with double-stranded DNA?
a) Fingers domain
b) Palm domain
c) Thumb domain
d) Exonuclease domain
[expand title=”Show answer” swaptitle=”Hide answer”] c) Thumb domain [/expand]
DNA polymerase I is most similar to which eukaryotic DNA polymerase in terms of its function?
a) DNA polymerase α
b) DNA polymerase β
c) DNA polymerase γ
d) DNA polymerase δ
[expand title=”Show answer” swaptitle=”Hide answer”] b) DNA polymerase β [/expand]
FAQ
What is DNA polymerase?
DNA polymerase is an enzyme responsible for catalyzing the synthesis of DNA molecules from deoxyribonucleotide triphosphates (dNTPs) during DNA replication.
Why is DNA polymerase important in biology?
DNA polymerase is crucial because it ensures the faithful replication of DNA, which is essential for passing genetic information from one generation to the next.
How does DNA polymerase work?
DNA polymerase works by adding complementary nucleotides to a template DNA strand, creating a new DNA strand with a sequence complementary to the template.
What is the role of DNA polymerase in DNA repair?
DNA polymerase is involved in DNA repair processes by replacing damaged or incorrect nucleotides with correct ones, thus maintaining genomic integrity.
What are the different types of DNA polymerases?
There are several DNA polymerases in both prokaryotes and eukaryotes, each with specific functions. Some examples include DNA polymerase I, DNA polymerase III, and DNA polymerase α.
Can DNA polymerase correct mistakes during DNA replication?
Yes, DNA polymerase has a proofreading function that allows it to correct errors by removing and replacing incorrect nucleotides.
What is the direction of DNA synthesis by DNA polymerase?
DNA polymerase synthesizes DNA in the 5′ to 3′ direction, meaning it adds new nucleotides to the 3′ end of the growing DNA strand.
Why is DNA polymerase I called a “processive enzyme”?
DNA polymerase I is considered processive because it can catalyze multiple polymerization steps without releasing the template strand.
What is the difference between DNA polymerase I and DNA polymerase III in E. coli?
DNA polymerase III is the primary replicative polymerase in E. coli, while DNA polymerase I primarily functions in DNA repair and removal of RNA primers during replication.
Can DNA polymerase I initiate DNA replication?
No, DNA polymerase I cannot initiate DNA replication. Replication is initiated by primase, which synthesizes a short RNA primer that DNA polymerase can extend from.
References
- Bhagavan, N. V., & Ha, C.-E. (2015). DNA Replication, Repair, and Mutagenesis. Essentials of Medical Biochemistry, 401–417. doi:10.1016/b978-0-12-416687-5.00022-1
- https://www.neb.com/en/products/m0209-dna-polymerase-i-e-coli#ProductInformation
- https://www.takarabio.com/products/cloning/modifying-enzymes/dna-polymerases/dna-polymerase-i-(pol-i)
- https://www.uniprot.org/uniprotkb/P00582/entry
- https://www.nzytech.com/en/mb42001-dna-polymerase-i-i-e-coli-i/
- https://www.biologyonline.com/dictionary/dna-polymerase-i