What is DNA Ladder?
- A DNA ladder is a solution composed of DNA molecules of varying lengths, which is predominantly utilized in gel electrophoresis techniques such as agarose or acrylamide gel electrophoresis. It is used to estimate the size of unidentified DNA fragments by comparing their mobility in an electrical field through the gel.
- DNA ladders, also known as molecular-weight size markers, allow researchers to distinguish and identify different DNA fragments based on their molecular weight, which correlates with their size. In numerous DNA-related procedures, such as delineating between DNA variants and determining the number of mutations present in a DNA fragment, they play a crucial role.
- Typically, DNA ladders are stained with nuclear stains such as ethidium bromide, enabling the visualization of DNA fragments following gel electrophoresis. DNA ladders sold commercially typically come in standard sizes of 50 base pairs (bp), 100 bp, 1000 bp, and 3000 bp.
- Using restriction enzymes, these DNA ladders are constructed by digesting DNA fragments of known lengths obtained from natural sources. The procedure is somewhat uncontrollable because the length of the fragments in the ladder is dependent on the restriction enzyme used.
- DNA engineering techniques have been devised to address this limitation and improve the adaptability of DNA ladders. A DNA fragment comprising a tandem repeat unit separated by the same unique restriction enzyme sites is cloned into a plasmid for commercial purposes. This plasmid is then partially digested to produce a ladder with multimers of the repeats, resulting in a ladder with greater versatility.
- Recently, laboratory protocols for preparing DNA ladders via polymerase chain reaction (PCR) have been developed. Either a single DNA target is amplified simultaneously using primer sets, or multiple DNA targets are amplified separately using specific primers. The PCR-based method provides greater control over the DNA ladder preparation procedure and permits the modification of ladder lengths to meet specific experimental requirements.
- In conclusion, DNA ladders are indispensable instruments in DNA laboratories for estimating the size of unidentified DNA fragments. They are molecular-weight size markers that help distinguish DNA fragments based on their molecular weight, and they are typically visualized with nuclear stains. DNA ladders can be generated through the digestion of known-length DNA fragments, DNA engineering techniques, and PCR-based techniques.
Definition of DNA Ladder
A DNA ladder is a solution containing DNA molecules of known lengths that are used as a reference to estimate the size of unknown DNA fragments separated through gel electrophoresis.
Characteristics of DNA Ladder
DNA ladders used for electrophoresis possess several important characteristics to ensure their effectiveness and reliability:
- Separability: The fragments within the DNA ladder should be distinct and separable from each other on the gel. This allows for accurate size estimation of unknown DNA fragments by comparing their migration distances relative to the ladder fragments.
- Visibility: The concentration of individual fragments in the DNA ladder should be sufficient to ensure their visualization after electrophoresis. This enables researchers to clearly observe and measure the ladder fragments alongside the unknown DNA fragments.
- Compatibility with loading dye: DNA ladders are often mixed with loading dye for easy loading onto the gel. It is crucial that the presence of the loading dye does not interfere with the specificity and accuracy of the DNA ladder, ensuring that the ladder fragments and unknown DNA fragments are not affected differently.
- Stability: The fragments in the DNA ladder should be stable over time, allowing for long-term storage and repeated use. This ensures consistent and reliable results when using the ladder as a size reference in various experiments.
- Purity: A high level of purification is essential for DNA ladders to avoid the presence of unnecessary or unknown fragments. Contaminants in the ladder can interfere with accurate size estimation and compromise the reliability of the ladder as a reference. Therefore, thorough purification processes are employed to eliminate unwanted fragments, ensuring the ladder consists solely of known-length DNA fragments.
By possessing these characteristics, DNA ladders provide researchers with a reliable size reference for estimating the size of unknown DNA fragments in gel electrophoresis, facilitating a wide range of DNA-related analyses and experiments.
Types of DNA ladder
- Standard DNA Ladders: These ladders consist of DNA fragments ranging from 100 base pairs (bp) to 10 kilobases (kb). They are commonly used for general DNA analysis, including size estimation and comparison of DNA fragments from different samples.
- High-Resolution DNA Ladders: High-resolution DNA ladders comprise DNA fragments ranging from 10 bp to 1 kb. They are used in applications that require precise size determination, such as genotyping and DNA sequencing.
- Denatured DNA Ladders: Denatured DNA ladders consist of DNA fragments that have been separated into individual strands. They are employed in applications like DNA fingerprinting and paternity testing.
- Quantitative DNA Ladders: Quantitative DNA ladders contain DNA fragments with known concentrations. They are used in DNA quantitation and quantitative polymerase chain reaction (qPCR) experiments.
- Low Molecular Weight (LMW) Ladders: LMW ladders consist of DNA fragments that are all shorter than 1,000 bp. They are suitable for analyzing DNA samples containing many small fragments, such as damaged cell DNA.
- High Molecular Weight (HMW) Ladders: HMW ladders consist of DNA fragments that are all longer than 1,000 bp. They are useful for analyzing DNA samples containing large fragments, such as DNA from chromosomes.
The choice of DNA ladder depends on the specific experimental requirements, including the desired size range of DNA fragments, resolution needs, and the application being pursued. Selecting the appropriate DNA ladder ensures accurate size estimation and reliable results in DNA analysis and molecular biology research.
There are various types of DNA ladders available from Jena Bioscience, each designed for specific applications and range of DNA fragment sizes. Here are the different types of DNA ladders:
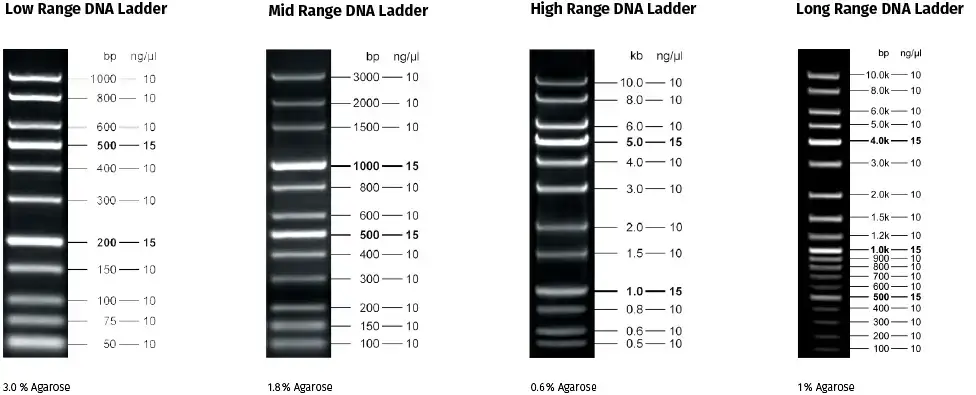
- Log Scale DNA Ladders:
- Log Scale DNA Ladders are ready-to-use and cover the full range of 20 bp to 10 kb. They provide size determination and concentration estimates of DNA fragments derived from PCR and restriction digests on agarose gels. Each fragment in this ladder is present at either 10 or 15 ng.
- Linear Scale DNA Ladders:
- Low Range DNA Ladder: This ladder covers a specific range of fragment sizes appropriate for low molecular weight DNA fragments.
- Mid Range DNA Ladder: This ladder covers a range of fragment sizes suitable for medium molecular weight DNA fragments.
- High Range DNA Ladder: This ladder covers a range of fragment sizes appropriate for high molecular weight DNA fragments.
- Long Range DNA Ladder: This ladder covers an extended range of fragment sizes, including both low and high molecular weight DNA fragments.
- Fluorescent Log Scale DNA Ladders:
- Fluorescent Log Scale DNA Ladders are similar to the Log Scale DNA Ladders mentioned earlier but are labeled with a fluorescent dye. They allow for visualization and quantification of DNA fragments using fluorescent detection methods.
- Fluorescent Linear Scale DNA Ladders:
- Fluorescent Linear Scale DNA Ladders are similar to the Linear Scale DNA Ladders but are labeled with a fluorescent dye. They provide accurate sizing and quantification of DNA fragments using fluorescent detection techniques.
- Classic DNA Ladders:
- λDNA/ HindIII Digest: This ladder consists of DNA fragments generated by digesting lambda DNA with the restriction enzyme HindIII.
- λDNA/ EcoRI Digest: This ladder consists of DNA fragments generated by digesting lambda DNA with the restriction enzyme EcoRI.
- λDNA/ EcoRI/ HindIII Digest: This ladder consists of DNA fragments generated by double digestion of lambda DNA with the restriction enzymes EcoRI and HindIII.
- λDNA/ StyI Digest: This ladder consists of DNA fragments generated by digesting lambda DNA with the restriction enzyme StyI.
- λDNA/ PstI Digest: This ladder consists of DNA fragments generated by digesting lambda DNA with the restriction enzyme PstI.
- λDNA/ BstEII Digest: This ladder consists of DNA fragments generated by digesting lambda DNA with the restriction enzyme BstEII.
These different types of DNA ladders offer researchers a range of options for accurate size determination, concentration estimation, and fragment visualization during gel electrophoresis, catering to specific experimental requirements.


How are DNA ladders made?
DNA ladders are typically generated through two main methods: enzymatic digestion and PCR amplification.
- Enzymatic Digestion Method:
- In this method, a known DNA template is fragmented using restriction enzymes. Restriction enzymes are enzymes that recognize specific DNA sequences and cut the DNA at those sites.
- The DNA template used can be either plasmids or PCR products. Plasmids are circular DNA molecules commonly used in molecular biology experiments.
- By selecting specific restriction enzymes, DNA fragments of different lengths are produced.
- The resulting DNA fragments are separated using gel electrophoresis to confirm their sizes and ensure the ladder’s integrity and purity.
- Once validated, the DNA fragments are mixed in specific ratios to create a ladder with known fragment sizes.
- PCR Amplification Method:
- In this method, DNA fragments of known lengths are generated through PCR (polymerase chain reaction) amplification.
- PCR is a technique that uses DNA polymerase enzyme to selectively amplify specific DNA sequences in a test tube.
- Primers, short DNA sequences that flank the target region, are designed to anneal to the desired DNA fragments.
- The PCR reaction is set up with the DNA template, primers, nucleotides, and DNA polymerase. Through a series of temperature cycles, the target DNA sequences are amplified.
- The amplified DNA fragments are separated using gel electrophoresis to confirm their sizes and purify the ladder.
- The resulting PCR products of known lengths are mixed in specific ratios to create a ladder with defined fragment sizes.
Both methods can be used to produce DNA ladders with different size ranges, depending on the desired application. Commercially available DNA ladders are often produced using a combination of these methods and may include additional purification steps to ensure high quality and accurate fragment sizes.
Examples of DNA ladder
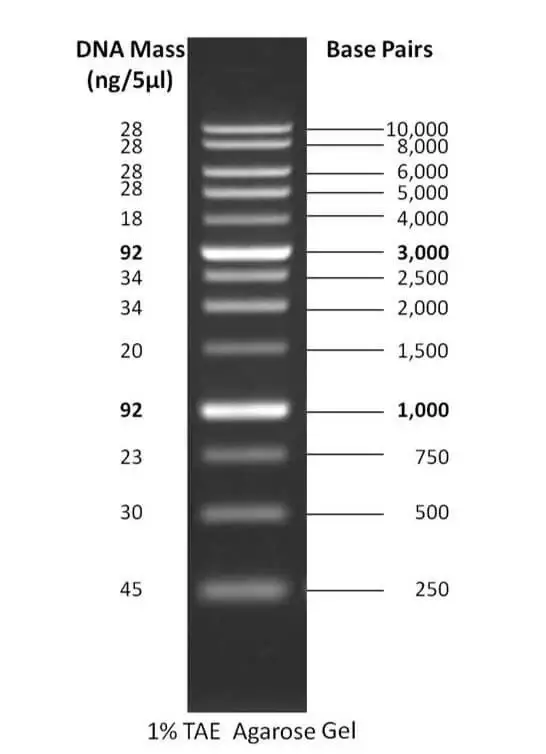
1. 1 kb DNA ladder
- The 1 kb DNA ladder is a commonly used molecular weight standard in molecular biology research. It is composed of 13 linear double-stranded DNA fragments, which enable the determination of the size of DNA fragments ranging from 250 base pairs (bp) to 10,000 bp.
- This ladder is created by combining a set of plasmids that have been digested with restriction enzymes and PCR products, resulting in a range of DNA fragments suitable for use as molecular weight markers in gel electrophoresis.
- The ladder is designed with reference bands at 1000 bp and 3000 bp, facilitating easy orientation during analysis. It is commercially available at various concentrations, but the recommended load for electrophoresis is typically 0.5 µg (5 µl).
- The 1 kb DNA ladder can be used in both agarose and polyacrylamide gels, with gel concentrations ranging from 0.75% to 1%. It comes with tracking dyes such as bromophenol blue and xylene cyanol FF to visualize the migration of the ladder fragments.
- For usage, the commercially available ladder is typically diluted in a 1:4 ratio with water, where 25 µl of the ladder is mixed with 75 µl of water. Loading dye provided with the kit is then added, and the solution is divided into 60 µl portions, which are stored at -20°C in microcentrifuge tubes.
- During gel electrophoresis, 3 µl of the diluted ladder solution is loaded per lane on a small agarose gel. This results in an approximate concentration of 0.63 µg of ladder DNA per lane.
- The 1 kb DNA ladder can be stored for different durations and at various temperatures, such as six months at 25ºC, 12 months at 4ºC, and 24 months at -20ºC. Appropriate storage buffers like 10 mM Tris-HCl or 1 mM EDTA are recommended.
- The main purpose of the 1 kb DNA ladder is to determine the size of double-stranded DNA fragments ranging from 250 bps to 10,000 bps. This allows for the quantification of DNA fragments and the comparison of their relative sizes. Additionally, the ladder serves as a control during the electrophoresis process, ensuring accurate size estimation of other DNA samples.
Applications
- Size Determination: The 1 kb DNA ladder is primarily used to determine the size of double-stranded DNA fragments ranging from 250 base pairs (bp) to 10,000 bp. By comparing the migration of DNA fragments in the ladder to the unknown samples, researchers can estimate the size of their target DNA fragments.
- Quantification of DNA Fragments: The ladder allows for the quantification of DNA fragments by providing a standard for comparison. Researchers can determine the relative abundance of DNA fragments in their samples based on their size compared to the ladder.
- Quality Control: The 1 kb DNA ladder serves as a control during the electrophoresis process. By running the ladder alongside the samples, researchers can verify the quality of the gel and the effectiveness of the electrophoresis run. Any inconsistencies or anomalies in the ladder bands may indicate issues with the gel or the experimental procedure.
- Standardization: The ladder acts as a molecular weight standard, enabling standardization across different experiments and laboratories. Researchers can compare their results to those obtained using the same ladder, facilitating the sharing and replication of research findings.
The 1 kb DNA ladder provides a convenient and reliable tool for size estimation, quantification, and quality assessment in gel electrophoresis experiments. Its versatility and widespread use make it an essential component in molecular biology laboratories for various applications involving DNA fragment analysis.
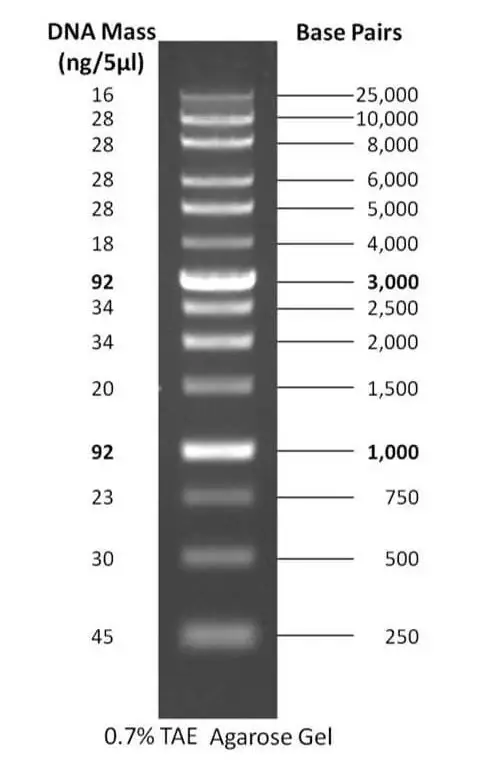
2. 1 kb plus DNA ladder
- The 1 kb plus DNA ladder is a DNA size standard commonly used in molecular biology research. It is composed of DNA fragments ranging in length from 0.5 kilobases (kb) to 10 kb, allowing for the determination of the size of double-stranded DNA fragments within the range of 250 base pairs (bp) to 25,000 bp.
- The 1 kb plus DNA ladder consists of 14 individual DNA fragments that have been purified using chromatography techniques. The purification process ensures the production of sharp and clear bands during electrophoresis, facilitating accurate size determination and identification of DNA fragments.
- These ladders typically contain 14 purified DNA fragments, including a reference band at 1000 bp and 3000 bp for easy orientation. The ladder may come pre-mixed with loading dye, which aids in the visualization of the ladder bands after electrophoresis.
- The 1 kb plus DNA ladder is precisely manufactured, ensuring the exact amount of DNA or number of DNA fragments in the ladder, allowing for accurate size estimation and consistent results.
- These ladders can be used in agarose gels or polyacrylamide gels with concentrations ranging from 0.7% to 1%.
- Overall, the 1 kb plus DNA ladder serves as a reliable molecular weight standard, providing researchers with a tool to determine the size of DNA fragments within a broad range. Its sharp bands, precise composition, and compatibility with various gel types make it a valuable asset in molecular biology experiments and DNA fragment analysis.
Uses of 1 kb plus DNA ladder
The 1 kb plus DNA ladder has several important uses in molecular biology research. Here are some key applications:
- Size Determination: The primary use of the 1 kb plus DNA ladder is to determine the size of double-stranded DNA fragments within a wide range, spanning from 250 base pairs (bp) to 25,000 bp. By comparing the migration of DNA fragments in the ladder to the unknown samples, researchers can estimate the size of their target DNA fragments.
- Quantification of DNA Fragments: The ladder enables the quantification of DNA fragments by providing a standard for comparison. Researchers can determine the relative abundance of DNA fragments in their samples based on their size compared to the ladder. This information is valuable for various applications, such as analyzing gene expression levels or assessing the efficiency of DNA manipulation techniques.
- Size Range Extension: The 1 kb plus DNA ladder allows for the determination of the size of DNA fragments that are larger than the typical range covered by regular 1 kb ladders. This expanded size range, reaching up to 25,000 bp, provides researchers with the ability to analyze and quantify longer DNA fragments, which may be important in certain studies and experiments.
- Quality Control: Similar to other DNA ladders, the 1 kb plus DNA ladder serves as a control during the electrophoresis process. Running the ladder alongside the samples allows researchers to assess the quality of the gel, confirm the proper function of the electrophoresis system, and verify the accuracy of their experimental setup.
- Standardization: The 1 kb plus DNA ladder acts as a molecular weight standard, facilitating standardization across experiments and laboratories. By using the same ladder, researchers can compare their results, reproduce experiments, and share findings more effectively.
3. 100 bp DNA ladder
- The 100 bp DNA ladder is a crucial tool in molecular biology laboratories used for the accurate sizing and quantification of double-stranded DNA fragments within the range of 100 base pairs (bp) to 1500 bp. It serves as a reference standard, providing a set of known DNA fragments with distinct sizes, allowing researchers to compare and identify the size of their DNA samples.
- The ladder consists of approximately 11 carefully purified DNA fragments, each having a specific size. These fragments are designed to form separate, easily distinguishable bands on an agarose gel when subjected to electrophoresis. By comparing the migration distance of an unknown DNA fragment with the ladder’s bands, scientists can determine the size of the unknown fragment.
- To facilitate the interpretation of results, the 100 bp DNA ladder typically includes reference bands at 500 bp and 1500 bp. These bands act as landmarks, aiding researchers in orienting themselves and identifying the approximate size of DNA fragments. The ladder is deliberately designed to have a uniform intensity of DNA bands across different sizes. This uniformity ensures that each band is clearly visible, allowing for accurate size determination.
- Quantification of DNA samples can also be estimated using the 100 bp DNA ladder. The ladder is prepared with a precise amount of DNA in each band, enabling researchers to make approximate measurements of the DNA concentration in their samples. This feature is particularly useful when determining the yield of PCR amplifications or the amount of DNA recovered from purification processes.
- In some cases, the 100 bp DNA ladder can be radio-labeled using enzymes like T4 polynucleotide kinase or T4 DNA polymerase. The radio-labeling allows for the visualization of the ladder bands through autoradiography, enhancing the sensitivity of detection.
- To visualize the ladder and DNA fragments, the 100 bp DNA ladder is typically run on 1-2% agarose gels. After electrophoresis, the gel is stained with a DNA-specific dye, such as ethidium bromide or SYBR Safe, which binds to the DNA and fluoresces under UV light. This fluorescence enables the clear visualization and documentation of the ladder’s bands, as well as any other DNA fragments present in the samples.
- In summary, the 100 bp DNA ladder is an indispensable tool in molecular biology laboratories. It serves as a reliable reference standard, allowing for the accurate determination of the size and approximate quantification of DNA fragments within a specific size range. With its distinct bands and reference markers, the ladder provides researchers with a valuable tool for analyzing and interpreting their DNA samples.

Applications of 100 bp DNA ladder
- Size Determination: The ladder is primarily employed for the accurate sizing of double-stranded DNA fragments within the range of 100 bp to 1500 bp. By comparing the migration distance of unknown DNA fragments to the ladder’s bands on an agarose gel, scientists can determine the size of their samples. This information is crucial for various downstream applications, such as cloning, sequencing, and genetic analysis.
- Diagnostic Purposes: The 100 bp DNA ladder plays a significant role in molecular diagnostics. It can be utilized in the molecular detection of pathogens, such as viruses or bacteria. By running patient samples alongside the ladder, researchers can identify the presence or absence of specific DNA fragments associated with the pathogen. This enables the diagnosis and characterization of infections, aiding in the development of appropriate treatments and control measures.
- Genetic Variability: Understanding the genetic variability within populations is essential in fields like evolutionary biology and population genetics. The 100 bp DNA ladder can be employed to characterize this genetic variability. By comparing the banding patterns of different individuals or populations on the ladder, researchers can analyze variations in DNA fragment sizes. This information helps to assess genetic diversity, relatedness, and evolutionary relationships between organisms.
- Molecular Tagging of Disease Resistance Genes: Disease resistance genes are vital in agriculture for developing crops with enhanced resistance to pathogens. The 100 bp DNA ladder can be utilized in the process of molecular tagging of disease resistance genes. By analyzing the association of specific DNA fragments from the ladder with resistance traits, researchers can identify and tag the genes responsible for resistance. This knowledge enables the breeding of crops with improved disease resistance, leading to higher crop yields and reduced reliance on chemical pesticides.
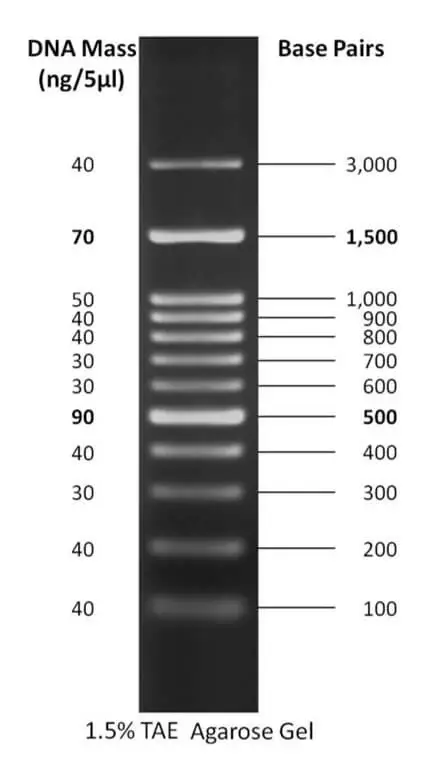
4. 100 bp plus DNA ladder
- The 100 bp plus DNA ladder is a versatile DNA size standard commonly used in molecular biology laboratories for the accurate sizing and quantification of double-stranded DNA fragments within the range of 100 base pairs (bp) to 3,000 bp. It serves as a reference tool, providing a set of known DNA fragments with distinct sizes, allowing researchers to compare and identify the size of their DNA samples.
- The ladder consists of approximately 12 highly purified DNA fragments, each carefully selected to cover a range of sizes within the desired range. These fragments are designed to form separate, easily distinguishable bands when subjected to electrophoresis on agarose or polyacrylamide gels. The clear bands facilitate the identification of DNA fragments within the same size range as the ladder, aiding researchers in determining the size of their unknown DNA fragments.
- To enhance ease of use and interpretation, the 100 bp plus DNA ladder includes reference bands at 500 bp and 1500 bp. These reference bands act as landmarks, providing a visual guide for orientation and approximate size estimation of DNA fragments. By comparing the migration distance of an unknown DNA fragment to the reference bands, researchers can quickly determine the size of the fragment.
- The ladder is designed to be visualized on 1-2% agarose or polyacrylamide gels. These gel concentrations are commonly used for DNA electrophoresis and provide optimal separation of fragments within the ladder’s size range. The ladder is typically run alongside the samples of interest, allowing for a direct comparison of fragment sizes.
- Each band within the ladder contains a defined amount of DNA, enabling researchers to make approximate quantification of their DNA samples. By comparing the intensity of the bands in their samples to the ladder’s bands, researchers can estimate the amount of DNA present in their samples. This feature is particularly useful when determining the yield of PCR amplifications or the amount of DNA recovered from purification processes.
- The 100 bp plus DNA ladder is generated through PCR amplification and restriction enzyme digestion of double-stranded DNA. The PCR amplification step ensures the production of sufficient quantities of DNA fragments of various sizes. Subsequent digestion with restriction enzymes further refines the ladder’s composition, providing a precise range of DNA fragment sizes.
- The ladder is supplied with a loading dye and loading buffer, which facilitate the movement of the ladder’s fragments through the gel slab during electrophoresis. The loading dye adds color to the DNA samples, making them visible during loading and ensuring efficient migration through the gel.
- In summary, the 100 bp plus DNA ladder is a valuable tool in molecular biology research. It allows for the accurate sizing and quantification of DNA fragments within a broad size range. With its clear bands, reference markers, and defined DNA amounts, the ladder provides researchers with a reliable reference standard for analyzing and interpreting their DNA samples on agarose or polyacrylamide gels.
Applications of 100 bp plus DNA ladder
The 100 bp plus DNA ladder serves several important purposes in molecular biology and genetic research. Here are some key uses of this DNA ladder:
- DNA Size Standard: The 100 bp plus DNA ladder is employed as a size standard for the visualization and determination of the size range of linear double-stranded DNA fragments. By running the ladder alongside DNA samples on an agarose or polyacrylamide gel, researchers can compare the migration distances of their samples to the ladder’s bands. This allows for the estimation of the molecular weight and size of the DNA fragments of interest.
- Identification and Separation: The ladder facilitates the identification and separation of DNA samples based on their molecular weight and size. By analyzing the migration patterns of the ladder’s bands alongside the samples, researchers can identify and distinguish different DNA fragments within the 100 bp to 3,000 bp range. This is particularly useful in applications such as cloning, sequencing, and genetic analysis, where accurate size determination is essential.
- Diagnostic Applications: The 100 bp plus DNA ladder also finds utility in diagnostic purposes, particularly in the molecular detection of pathogens. By running patient samples alongside the ladder, researchers can detect and identify specific DNA fragments associated with the presence of pathogens. This enables the diagnosis and characterization of infections, aiding in the development of targeted treatments and control strategies. The ladder serves as a reference point for the identification of pathogen-specific DNA fragments in diagnostic assays.
- Quality Control: The ladder is crucial for quality control in molecular biology experiments. It serves as a benchmark to assess the performance of gel electrophoresis systems, ensuring the accuracy and reproducibility of DNA fragment size determination. By comparing the migration patterns of the ladder’s bands to the expected sizes, researchers can verify the integrity and efficiency of their experimental setup.
- Teaching and Training: The 100 bp plus DNA ladder is widely used in educational settings for teaching and training purposes. It serves as a visual aid in demonstrating the principles of gel electrophoresis and DNA fragment analysis. By visualizing and interpreting the ladder’s bands, students can learn about DNA size determination, molecular weight estimation, and the significance of reference standards in molecular biology experiments.
Advantages of DNA Ladders
- Size reference: DNA ladders provide a known size reference for DNA fragments. They allow researchers to determine the approximate size of their DNA samples by comparing their migration on a gel to the ladder fragments.
- Easy identification: DNA ladders typically have reference bands at specific sizes, which make it easy to identify and assign sizes to other DNA fragments in the gel. This aids in the interpretation and analysis of experimental results.
- Quantification: DNA ladders can be used for rough quantification of DNA samples. By comparing the intensity of unknown DNA bands to ladder bands of known concentration, researchers can estimate the amount of DNA present in their samples.
- Quality control: DNA ladders serve as a quality control tool in gel electrophoresis experiments. Their presence in each gel run helps researchers assess the integrity of their DNA samples and ensure that the experiment is functioning correctly.
- Versatility: DNA ladders come in various sizes and configurations, catering to different research needs. They can be used in different types of electrophoresis gels, including agarose and polyacrylamide gels.
Limitations of DNA Ladders
- Limited size range: Each DNA ladder has a specific size range, and it may not cover all possible DNA fragment sizes of interest. Researchers may need to use different ladders or combine multiple ladders to cover a broader range of fragment sizes.
- Resolution limitations: The resolution of DNA ladders may not be sufficient to distinguish very close fragment sizes. This can make it challenging to accurately determine the size of DNA fragments that fall within a similar size range.
- Controllable size variation: DNA ladders produced through digestion by restriction enzymes may have limited control over the precise sizes of ladder fragments. This can result in slight variations in fragment lengths between different batches or manufacturers.
- Cost: DNA ladders can be relatively expensive, especially for high-resolution or specialized ladders. This cost may pose a limitation for researchers working on a tight budget or conducting large-scale experiments.
- Limited information: DNA ladders provide information about size but not sequence or composition. They do not offer insights into the specific genetic content or sequence variations present in the DNA fragments.
Applications of DNA Ladders
DNA ladders have several important applications in molecular biology and genetic research. Here are some common applications of DNA ladders:
- Size determination: DNA ladders are primarily used to determine the size of unknown DNA fragments by comparing their migration distance on a gel to the known lengths of ladder fragments. This is essential for accurate sizing of DNA fragments in various experiments.
- Quantification: DNA ladders can provide an approximate estimation of DNA concentration by comparing the intensity of unknown DNA bands to the ladder bands. This can be helpful in determining the amount of DNA present in a sample.
- Quality control: DNA ladders serve as a quality control measure in gel electrophoresis experiments. By including a ladder in each gel run, researchers can assess the integrity of their DNA samples and ensure that the experiment is functioning properly.
- Gel visualization: DNA ladders are often stained with DNA-binding dyes, such as ethidium bromide or SYBR Green, to visualize the DNA fragments after gel electrophoresis. This allows researchers to confirm the presence and distribution of DNA fragments in the gel.
- Molecular weight determination: DNA ladders, also known as molecular weight markers, aid in the determination of the molecular weight or size of DNA fragments. This information is crucial for understanding the structure and function of DNA molecules.
- Fragment separation optimization: DNA ladders can be used to optimize the separation conditions in gel electrophoresis experiments. By analyzing the migration patterns of the ladder fragments under different conditions, researchers can fine-tune parameters such as gel concentration, buffer composition, and voltage for optimal resolution of DNA fragments.
- PCR product size verification: DNA ladders can be used to verify the size of PCR products. By comparing the migration distance of PCR products to the ladder fragments on a gel, researchers can confirm the expected size of the amplified DNA fragments.
- DNA sequencing: DNA ladders with high-resolution fragments are often used as size standards in DNA sequencing experiments. They help in determining the precise size of DNA fragments generated during the sequencing process, which is crucial for accurate sequence analysis.
- DNA fingerprinting and genotyping: DNA ladders are utilized in DNA fingerprinting and genotyping techniques, such as restriction fragment length polymorphism (RFLP) analysis. They provide reference points for comparing the sizes of DNA fragments generated by different individuals or samples.
- Teaching and training: DNA ladders are widely used in educational settings, such as biology labs and training courses, to demonstrate principles of DNA analysis, gel electrophoresis, and fragment sizing. They serve as a valuable tool for teaching basic molecular biology techniques and concepts.
DNA Ladders Concept Map/Cheatsheet

FAQ
What is a DNA ladder?
A DNA ladder is a solution of DNA fragments of known lengths used as a reference to determine the size of unknown DNA fragments during gel electrophoresis.
What is the purpose of using a DNA ladder?
The main purpose of using a DNA ladder is to estimate the size of unknown DNA fragments by comparing their migration distance on a gel to the known lengths of the ladder fragments.
How are DNA ladders made?
DNA ladders can be produced by digesting known-length DNA fragments from natural sources using restriction enzymes. Alternatively, DNA engineering techniques or PCR amplification methods can be employed to create ladder fragments.
What are the common sizes available in commercial DNA ladders?
Commercial DNA ladders are available in various sizes, such as 50 bp, 100 bp, 1 kb, and 3 kb. However, there are other options available as well, depending on the specific requirements of the experiment.
Can DNA ladders be used with different types of gel electrophoresis?
Yes, DNA ladders can be used with different types of gel electrophoresis, including agarose gels and polyacrylamide gels. The choice of gel type depends on the size range of the DNA fragments being analyzed.
How are DNA ladders visualized?
DNA ladders are typically stained with DNA-binding dyes, such as ethidium bromide or SYBR Green, which allow the visualization of DNA fragments under ultraviolet (UV) light.
Are DNA ladders stable for long-term storage?
DNA ladders can be stored for extended periods if kept at appropriate temperatures (-20°C or below) and in suitable storage buffers to prevent degradation. The stability may vary depending on the specific ladder and storage conditions.
Can DNA ladders be used for DNA quantification?
DNA ladders can provide an approximate estimation of DNA concentration by comparing the intensity of unknown DNA bands to the ladder bands. However, for accurate DNA quantification, dedicated DNA quantitation methods should be employed.
Are there specialized DNA ladders for high-resolution applications?
Yes, there are high-resolution DNA ladders available that contain smaller DNA fragments, such as 10 bp to 1 kb. These ladders are used for applications that require fine resolution, such as DNA sequencing or genotyping.
Can DNA ladders be used for DNA fragment sizing in different organisms?
Yes, DNA ladders can be used for DNA fragment sizing in various organisms, as the ladder fragments are of known lengths and can be compared to the unknown DNA fragments irrespective of the source organism.
References
- https://www.thermofisher.com/in/en/home/life-science/dna-rna-purification-analysis/nucleic-acid-gel-electrophoresis/dna-ladders.html
- https://international.neb.com/products/markers-and-ladders/dna-markers-and-ladders/dna-markers-and-ladders
- https://goldbio.com/collection/dna-ladders
- https://sites.psu.edu/tanlab/dna-ladder-plasmids/
- https://www.gbiosciences.com/DNA-Ladders
- https://bayoubiolabs.com/ladders.html
- https://www.smobio.com/shop/category/dna-ladders-5?order=list_price+desc
- https://us.bioneer.com/products/laddersandmarkers/100bpDNALaddertechnical.aspx?AspxAutoDetectCookieSupport=1
- https://www.thermofisher.com/in/en/home/life-science/dna-rna-purification-analysis/nucleic-acid-gel-electrophoresis/dna-ladders.html
- https://www.jenabioscience.com/molecular-biology/dna-ladders