What is Helicase?
- Helicases are a class of enzymes that play a fundamental role in unwinding nucleic acids, either DNA or RNA, depending on the type of helicase involved. These enzymes are essential for various cellular processes, including replication, repair, transcription, and translation. Helicases were first identified in E. coli in 1976, and later, eukaryotic DNA helicases were discovered in 1978. Approximately 1% of the human genome encodes genes responsible for producing these helicases.
- There are two main types of helicases: DNA helicases and RNA helicases. In humans, 31 DNA helicases and 64 RNA helicases are encoded by different genes. One critical aspect of helicases is that they require energy in the form of ATP (adenosine triphosphate) to perform their functions, which involve breaking the hydrogen bonds between nucleotides and separating the strands of nucleic acids.
- Because the process of replication is universal to all living organisms, helicases are found across all forms of life, from simple viruses to complex eukaryotic cells. DNA helicase, in particular, is crucial for the replication process. It unwinds the DNA helix, allowing the replication machinery to copy the genetic material accurately. RNA helicases, on the other hand, are involved in the regulation of RNA metabolism, including destabilizing RNA secondary structures, assisting in RNA splicing, regulating RNA function, and contributing to ribosome assembly.
- Interestingly, some helicases are specialized to work on DNA-RNA hybrids. These DNA-RNA helicases separate the hybrid strands and play a role in terminating transcription, ensuring that cellular processes are tightly regulated.
- DNA helicase is particularly significant due to its role in DNA replication, disease development, and its use in biological experiments. It ensures that genetic information is accurately copied and transmitted during cell division. Malfunctions in helicase activity can lead to genetic instability and contribute to various diseases, including cancer and genetic disorders.
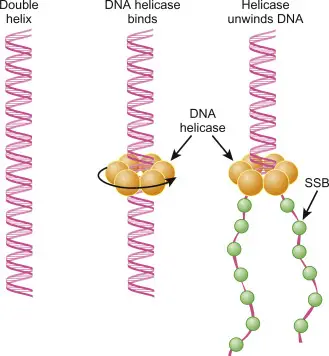
What is DNA Helicase?
- DNA helicase is a crucial enzyme that plays an essential role in various cellular processes involving DNA. Its primary function is to unwind the double-stranded DNA molecule, enabling the separation of two complementary strands. This separation is necessary for critical biological processes such as DNA replication, transcription, recombination, and repair. DNA helicases achieve this by breaking the hydrogen bonds between the nitrogenous base pairs, utilizing energy derived from the hydrolysis of ATP (adenosine triphosphate).
- DNA helicases are motor proteins that move directionally along the DNA phosphodiester backbone, ensuring that the strands of DNA can be properly separated. This activity generates what is known as a replication fork, a Y-shaped structure where the unwinding of the DNA occurs, providing a template for the synthesis of new DNA strands. The replication fork is a critical component in ensuring that DNA replication proceeds correctly, maintaining the integrity and accuracy of genetic information as it is passed from one cell generation to the next.
- In eukaryotic cells, the human genome encodes 31 distinct DNA helicases, reflecting the diverse and vital roles these enzymes play across different processes. They not only unwind double-stranded DNA but are also involved in resolving complex DNA structures such as triplexes or G-quadruplexes. Additionally, DNA helicases are capable of displacing proteins that bind to single or double-stranded DNA, facilitating the continuation of essential cellular processes.
- The mechanism by which helicases operate relies on three common biochemical properties: nucleic acid binding, NTP (nucleoside triphosphate) or dNTP (deoxynucleoside triphosphate) binding, and hydrolysis, followed by an ATP-dependent unwinding of nucleic acid strands. These enzymes can work in either the 3’ to 5’ or 5’ to 3’ direction, depending on the type of DNA helicase and its specific function.
- Discovered in Escherichia coli in 1976, DNA helicases are ubiquitous across all domains of life, highlighting their fundamental role in cellular survival and genome stability. Moreover, certain helicases have specialized functions beyond DNA replication and repair, including their involvement in immune responses where they sense viral nucleic acids.
- DNA helicases are also part of intricate systems that recover stalled replication forks. For example, enzymes like RecG and PriA help in the regression of DNA, creating a structure known as a Holliday junction, which is important in the repair and resumption of DNA replication after damage or replication stress.
History of DNA Helicases
The history of DNA helicases begins with their discovery in the mid-1970s. These enzymes, crucial for unwinding DNA strands, have been extensively studied across a wide range of organisms. From bacteria to plants and humans, research on helicases has revealed their essential role in DNA replication, repair, and recombination. Below is a timeline of key discoveries that mark significant milestones in the history of DNA helicase research:
- 1976: The first DNA helicase was discovered and isolated from Escherichia coli. Described as a “DNA unwinding enzyme,” it was characterized by its ability to denature DNA duplexes in an ATP-dependent reaction without degrading the strands. This was the foundational step in understanding the enzymatic activity of helicases in DNA metabolism.
- 1978: The first eukaryotic DNA helicase was discovered, isolated from the lily plant. This discovery demonstrated that helicases are not exclusive to prokaryotic systems but also play a critical role in eukaryotes.
- 1982: In bacteriophages, DNA helicases were identified with the discovery of the “T4 gene 41 protein” as the first reported bacteriophage DNA helicase. This further highlighted the role of helicases in viral replication.
- 1985: Mammalian DNA helicases were first isolated from calf thymus, marking the discovery of helicases in higher organisms. This was a significant step toward understanding helicase function in more complex eukaryotic systems.
- 1986: The SV40 large tumor antigen was reported as the first viral helicase. This finding revealed the dual functionality of certain viral proteins in both tumorigenesis and DNA unwinding during viral replication.
- 1986: In yeast, ATPaseIII was identified as a DNA helicase, broadening the scope of helicase function in lower eukaryotes and laying the groundwork for future studies in yeast as a model organism for helicase activity.
- 1988: The discovery of seven conserved amino acid domains, known as helicase motifs, provided molecular insight into the structural and functional aspects of helicases. These motifs became essential in the identification and classification of helicases across different organisms.
- 1989: Helicases were classified into Superfamily I and Superfamily II, establishing a system for organizing the growing number of helicases based on sequence and functional similarities.
- 1989: The DEAD box helicase family was identified, representing a large group of RNA helicases that participate in various aspects of RNA metabolism. This family is characterized by the presence of a conserved sequence motif that includes the amino acids Asp-Glu-Ala-Asp (DEAD).
- 1990: The first human DNA helicase was isolated, underscoring the relevance of helicases in human cellular processes, including DNA replication, repair, and transcription.
- 1992: The first mitochondrial DNA helicase was discovered in bovine brain tissue. This finding provided insight into the unique mechanisms of DNA maintenance in the mitochondria, which have their own distinct genome.
- 1996: The first purified chloroplast DNA helicase was reported in peas, adding to the understanding of DNA replication in plant organelles, particularly in the context of the chloroplast genome.
- 2002: A biochemically active DNA helicase was isolated from Plasmodium cynomolgi, a malarial parasite. This marked the first such discovery in a parasitic organism, offering potential targets for antimalarial drug development.
Biochemical Properties of Helicases
The following points detail the key properties and mechanisms of helicases, highlighting their importance in nucleic acid dynamics.
![DNA Helicases - Structure, Types, Mechanism, Functions, Examples 2 Biochemical Properties of Helicases: (a) Helicase (blue triangle) unwinds duplex DNA by hydrolyzing ATP, using the energy to separate the strands into single strands. (b) An ssDNA translocase (blue triangle) moves directionally along DNA, driven by ATP binding and hydrolysis, with translocation occurring in either the 3’–5’ or 5’–3’ direction. (c) A dsDNA translocase (red hexagon) advances along double-stranded DNA. (d) Helicases can displace proteins (magenta circles) bound to the DNA strand due to their directional movement (adapted from ref. [45]).](https://biologynotesonline.com/wp-content/uploads/2024/10/image-291.png)
- Nucleic Acid Binding:
- Helicases exhibit a strong preference for binding to single-stranded DNA (ssDNA) compared to double-stranded DNA (dsDNA). This property is essential for their functionality, as they often require a short stretch of ssDNA to initiate unwinding.
- The loading of helicases onto their nucleic acid substrates is often a rate-limiting step in the unwinding process, determining the efficiency of subsequent reactions.
- ATP Binding and Hydrolysis:
- The activity of helicases is tightly coupled to ATP binding and hydrolysis. ATP hydrolysis provides the energy necessary for helicases to translocate along nucleic acids and unwind duplex structures.
- Although the ATPase activity of helicases is low in the absence of nucleic acids, the presence of ssDNA significantly stimulates this activity, enhancing their unwinding capability.
- Directionality of Movement:
- Helicases move in a unidirectional manner, either in a 5′→3′ or 3′→5′ direction. This movement is determined by the polarity of the nucleic acid strand to which the helicase is bound.
- The inherent directionality impacts substrate specificity, as a 5′→3′ helicase may bind poorly to substrates with a 3′ ssDNA overhang but has a high affinity for those with a 5′ overhang. For example, the RecBCD helicase exhibits bipolar activity, with its RecB and RecD components unwinding DNA in opposite directions.
- Translocation Rate:
- The translocation rate of helicases varies significantly among different helicases, with some moving only a few base pairs per second, while others can translocate thousands of base pairs in the same time frame.
- This rate is generally governed by the hydrolysis of ATP, which powers the helicase’s movement along the nucleic acid substrate.
- Step Size:
- Step size refers to the number of base pairs unwound or translocated during one catalytic cycle of ATP hydrolysis. This characteristic is vital for understanding how efficiently a helicase can unwind nucleic acids.
- Different helicases exhibit varying step sizes, which directly influences their unwinding capability.
- Processivity:
- Processivity describes a helicase’s ability to catalyze multiple rounds of unwinding without dissociating from the substrate. Most helicases are highly processive, allowing them to efficiently unwind long stretches of DNA.
- The processivity of a helicase can be enhanced by interactions with partner proteins, which may stabilize the helicase-DNA complex and regulate its activity.
- Active vs. Passive Unwinding:
- Active unwinding occurs when a helicase physically separates the strands of DNA, a process that requires ATP hydrolysis. This function is crucial for numerous cellular processes, including replication and repair.
- Conversely, passive unwinding refers to the helicase’s binding to regions of DNA that are already unwound, often due to transient strand dissociation—a phenomenon known as “nucleic acid breathing.” In this case, the helicase does not actively separate the strands but rather stabilizes already unwound regions.
Structure of DNA Helicase
Below is a detailed exploration of the structure of DNA helicases, focusing on their key features and functional components.
- Hexameric Structure:
- Many DNA helicases form a ring-like hexameric structure, which consists of six subunits that surround the DNA. This ring structure creates a central channel through which the DNA strand passes as the enzyme works.
- The hexameric form is particularly efficient for stability and processivity, meaning the helicase can maintain prolonged interaction with the DNA and move along the strand without disengaging frequently. This topological link between the protein and nucleic acid enhances its effectiveness.
- While most hexameric helicases are homohexamers (composed of identical subunits), eukaryotic DNA helicases such as the minichromosome maintenance (MCM) helicases are an exception, as they contain non-identical subunits.
- Conserved Motifs:
- DNA helicases possess a set of conserved sequence motifs in their catalytic core, which are crucial for the enzyme’s activity. These motifs are often clustered in the central region of the protein, typically ranging from 200 to 700 amino acids.
- The most common motifs in DNA helicase include Walker A (Motif I) and Walker B (Motif II), which are essential for ATP binding and hydrolysis. Other motifs, such as Motifs III, IV, V, VI, and Motif Q, also contribute to the enzymatic function. These conserved sequences are responsible for interacting with ATP and the DNA molecule, facilitating the unwinding process.
- AAA+ Domain:
- A key feature of many helicases, particularly those with hexameric structures, is the presence of an AAA+ (ATPases Associated with diverse cellular Activities) domain. This domain functions as the nucleotide-binding site and is integral to the catalytic activity of the enzyme.
- The AAA+ domain is where the enzyme first interacts with DNA nucleotides, using the energy from ATP hydrolysis to break the hydrogen bonds between the DNA strands.
- Monomeric and Dimeric Forms:
- While hexameric helicases are common, other structural forms also exist. Some helicases are found as monomers or dimers, though these forms are less common than the hexameric variety. The structural variation allows helicases to function in different cellular environments or under different conditions.
- Ring and Non-Ring Helicases:
- Not all helicases form ring-like structures. There are helicases that do not adopt the hexameric ring shape, and these non-ring helicases function through different mechanisms of action. The ring-forming helicases are generally more processive and are better suited for long stretches of DNA or RNA, while non-ring helicases may have more specialized or localized roles.
Helicase Motifs
A defining characteristic of helicases is the presence of conserved amino acid sequences known as “helicase motifs.” These motifs not only facilitate the classification of helicases into distinct families but also provide insights into their functional mechanisms. The following points outline the key features and functions of helicase motifs, specifically focusing on the SF1 and SF2 helicase families.
- Conserved Amino Acid Motifs:
- Helicases can be grouped into families based on the sequence similarity and arrangement of their conserved motifs. Notably, the SF1 and SF2 families each possess at least seven conserved motifs, which exhibit significant similarities in their sequence and secondary structure.
- Within the SF1 family (including Rep and PcrA helicases) and the SF2 family (represented by NS3), motifs III and IV are key points of differentiation. For instance, in NS3, motif IV interacts with the DNA backbone differently compared to the same motif in Rep and PcrA helicases.
- Core and Divergent Regions:
- The conserved motifs are typically clustered within a core region of 200–700 amino acids. This core is interspersed with stretches that, while low in sequence conservation, are high in length conservation.
- Conversely, the N-terminal and C-terminal regions of helicases exhibit significant variability in sequence and length. These divergent regions are crucial for the unique functions of individual helicases, while the conserved motifs are predominantly involved in ATP binding and hydrolysis, as well as nucleic acid binding and unwinding.
- Functional Motifs:
- Q Motif: This motif is universally conserved among SF1 helicases. It plays a critical role in coordinating the adenine base of ATP and is essential for orienting ATP for hydrolysis.
- Motif I (Walker A): Characterized by the consensus sequence AxxGxGKT, this motif is found in various nucleotide-binding proteins. It forms a phosphate-binding loop, with the GKT residues being vital for interaction with Mg²⁺ and ATP.
- Motif Ia: The consensus sequence TxxAA suggests involvement in ssDNA binding and energy transduction from the ATP-binding site to the DNA-binding site. In SF1 helicases like UvrD, Rep, and Pif1, motifs Ia and III are crucial for determining the direction of translocation.
- Motif II (Walker B): Represented by the sequence DExx, this motif is pivotal for NTP hydrolysis. The D and E residues coordinate with Mg²⁺ and activate the attacking water molecule, thereby facilitating ATPase activity.
- Motif III: The consensus sequence GDxxQLPP indicates its role in DNA binding through base stacking and hydrogen bonding. Motif III is also implicated in transducing the energy derived from ATP hydrolysis to the DNA substrate, as evidenced by mutants displaying uncoupled ATPase and helicase activities.
- Motif IV: This motif varies among the SF1 helicase family but is generally involved in providing a platform for the adenine base and making direct contact with the γ-phosphate, suggesting its role in NTP hydrolysis.
- Motif V: This motif is responsible for interacting with the sugar-phosphate backbone of DNA. Mutations in motif V can lead to diminished ssDNA affinity and reduced ATP hydrolysis rates.
- Motif VI: Defined by the sequence VA(L/Y)TRA(K/R), this motif is part of the ATP-binding cleft and plays a crucial role in coupling the helicase and ATPase activities. Mutants of this motif often exhibit defects in nucleic acid binding and reduced rates of ATP hydrolysis.
- Functional Interactions:
- The conserved motifs collaborate to bind and hydrolyze ATP, which triggers conformational changes essential for helicase function. These changes drive the directional movement of helicases along single-stranded (ssDNA) or double-stranded (dsDNA) substrates.
- The motifs facilitate communication between the nucleic acid-binding sites and the ATP-binding sites, enabling efficient helicase activity.
Types of DNA Helicases
DNA helicases are essential enzymes involved in various cellular processes, and their structural and functional diversity reflects the wide range of roles they play in DNA metabolism. Based on their sequence similarities and conserved motifs, DNA helicases are categorized into six superfamilies (SF1 to SF6). Below is an organized breakdown of these superfamilies, their structures, and their functional roles:
- Superfamily SF1:
- SF1 helicases have a characteristic structure featuring two α-β RecA-like domains that play a central role in several cellular processes, including recombination, transcription, and repair.
- The structural core of SF1 helicases includes tandem alpha-helices and five contiguous parallel beta-strands, with ATP binding occurring at the amino-terminal α-β domain. This region contains the conserved motifs Walker A (Motif I) and Walker B (Motif II), essential for ATP binding and hydrolysis.
- SF1 helicases are further subdivided into three subfamilies based on function and translocation direction. These include the PiF1/RecD, Rep/UvrD, and UpF1-like subfamilies. SF1A translocates in the 3′ to 5′ direction on single-stranded DNA (ssDNA), while SF1B moves in the 5′ to 3′ direction.
- Superfamily SF2:
- SF2 helicases are primarily involved in RNA metabolism, though they also participate in several steps of DNA metabolism. This superfamily is more functionally diverse than SF1, with 10 distinct families of helicases found within SF2. Each family serves specialized roles in nucleic acid processing.
- Superfamily SF3:
- Helicases within SF3 are typically associated with small DNA and RNA viruses, as well as large nucleocytoplasmic DNA viruses. The SF3 helicases have a conserved structural arrangement where the Walker A and Walker B motifs, responsible for ATP hydrolysis, are separated by a spacer region. A third motif, Motif C, lies between Walker B and the C-terminus of the conserved region.
- Superfamily SF4:
- SF4 helicases are hexameric in structure, with a central core similar to the α-β RecA-like domain. This superfamily is primarily involved in bacterial and bacteriophage replication, closely related to bacterial DnaB protein.
- Superfamily SF5:
- A prominent example of SF5 helicases is the E. coli Rho factor, which functions in terminating specific RNA transcripts. The SF5 helicases are hexameric and translocate on RNA. The structure and function of SF5 helicases shed light on how replicative helicases operate in cells.
- Superfamily SF6:
- SF6 helicases contain the AAA+ core, which is absent in SF3 helicases. Members of this group include proteins involved in DNA replication and repair, such as minichromosome maintenance (MCM) helicases, RuvA, RuvB, and RuvC. These proteins are critical for ensuring the proper replication and maintenance of chromosomal integrity.
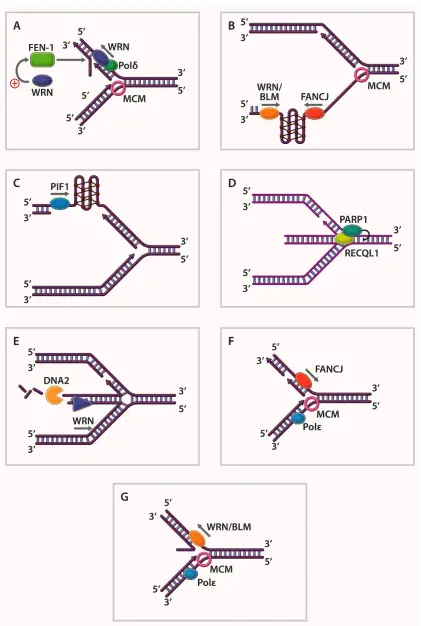
Mechanism of DNA Helicases
Below is a detailed explanation of how DNA helicases function:
- Helicase Activation: The process begins when the helicase recognizes and binds to a dsDNA strand. A short single-stranded DNA (ssDNA) region is required for the helicase to initiate the unwinding process. This typically happens in vitro, unlike the full-length dsDNA unwinding in natural replication.
- Oligomeric Assembly: DNA helicases often form oligomeric structures, with hexameric assemblies being the most common and effective for full activity. In this hexameric arrangement, six helicase subunits form a ring that encircles the DNA strand. The assembly is stabilized through the binding of nucleoside triphosphates (NTPs) or metal ions, essential for maintaining structural integrity during the unwinding process.
- ATP Binding and Hydrolysis: ATP binding sites are crucial for helicase activity. In a hexameric helicase, six potential ATP-binding sites exist, but only two opposite sites bind ATP tightly, two bind ADP and inorganic phosphate, and the remaining two remain empty. ATP hydrolysis provides the necessary energy for the helicase to overcome the activation barrier required to break hydrogen bonds between nucleotides.
- Conformational Changes: Upon ATP hydrolysis, a coordinated ripple effect occurs around the helicase ring. This causes conformational shifts, particularly in the loops extending toward the central channel of the helicase. These loops oscillate, dragging the DNA strand through the central pore of the helicase, leading to the unwinding of the DNA double helix.
- Nucleotide Binding and DNA Separation: The core of the helicase, often associated with the AAA+ domain, acts as the nucleotide-binding site where ATP binds and is hydrolyzed. This hydrolysis provides energy to break hydrogen bonds between the base pairs, resulting in strand separation. The DNA-binding motifs, such as Walker A and Walker B, play a crucial role in ATP hydrolysis and subsequent movement of the helicase along the DNA strand.
- Directional Movement: Helicases can operate in either a unidirectional or bidirectional manner. For instance, the RecB helicase functions unidirectionally, moving from the 3’ to 5’ end along the DNA strand. In contrast, ReBCD helicase exhibits bidirectional movement, translocating in both the 5’ to 3’ and 3’ to 5’ directions, depending on its specific role in DNA repair or replication.
- Processivity of Helicase: The continuous binding and hydrolysis of ATP enable the helicase to move forward one nucleotide at a time. The helicase progresses along the DNA strand, hydrolyzing ATP with each step and breaking the hydrogen bonds between base pairs, thereby separating the strands.
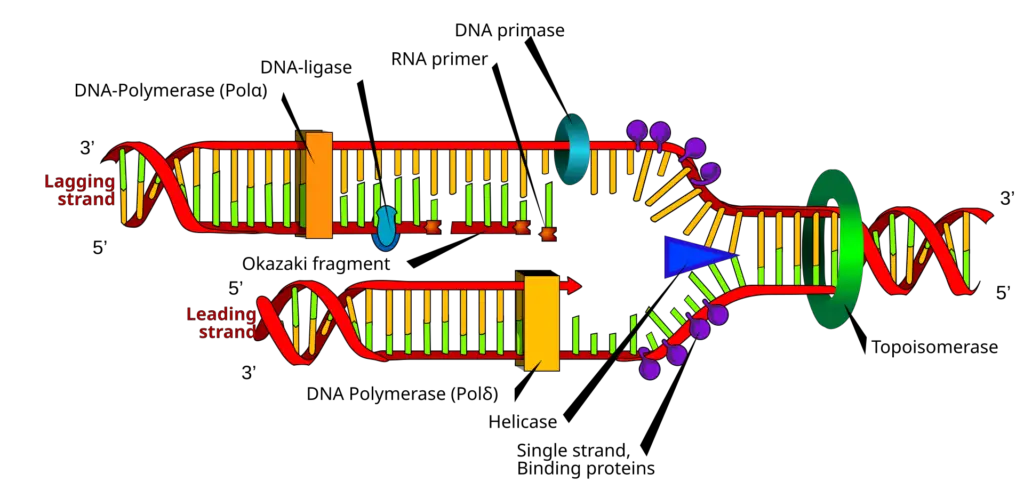
Prokaryotic DNA helicases
Prokaryotic DNA helicases are crucial enzymes that facilitate the unwinding of DNA, a fundamental process in various cellular functions such as replication, repair, and recombination. The study of these helicases in Escherichia coli has led to the identification of eleven distinct helicases, which can be broadly categorized into three functional groups: those involved in DNA replication, those involved in DNA repair and recombination, and other helicases with diverse functions.
- Replicative Helicases:
- DnaB Protein:
- This hexameric helicase, composed of identical 52 kDa monomers, is essential for DNA replication, particularly in unwinding duplex DNA at the replication fork.
- DnaB requires a 3′ single-stranded tail for effective interaction with DNA, demonstrating a preference for replication fork-like substrates.
- Its unwinding rate reaches approximately 730 nucleotides per second with a high processivity, capable of unwinding over 50,000 base pairs.
- DnaB initiates bidirectional unwinding at the origin of replication in collaboration with DnaC and DnaG primase, ensuring the synthesis of RNA primers for lagging strand synthesis.
- PriA Protein:
- PriA exhibits a unique ability to interact with DNA based on nucleotide sequence and is known to translocate in a 3′ to 5′ direction.
- It plays a pivotal role in the formation of the preprimosome complex, essential for the assembly of the primase.
- Genetic studies indicate that although cells lacking PriA can survive, they display significant growth defects, emphasizing its role in effective primosome assembly rather than direct helicase activity.
- Rep Protein:
- Required for the replication of several single-stranded DNA phages, Rep protein unwinds DNA through a catalytic mechanism, functioning efficiently in unwinding the replicative form of viral chromosomes.
- Mutations in the rep gene correlate with increased replication fork numbers and reduced movement rates, suggesting its involvement in maintaining efficient DNA replication.
- Helicase II (UvrD):
- This helicase unwinds duplex DNA using a concentration-dependent mechanism, and is notable for its role in DNA repair and recombination pathways.
- It is implicated in both excision repair and mismatch repair, with its activity being crucial for the turnover of the UvrC protein during DNA repair processes.
- Genetic evidence suggests that UvrD functions in recombination alongside RecE and RecQ helicases, indicating overlapping roles among helicases in different pathways.
- DnaB Protein:
- Repair and Recombination Helicases:
- UvrAB Complex:
- The UvrAB complex, composed of UvrA and UvrB, plays a role in nucleotide excision repair by recognizing and unwinding damaged DNA regions.
- The unwinding activity of this complex is restricted to short duplex segments, critical for subsequent repair processes.
- RecQ Helicase:
- This helicase contributes to the recombination pathway by generating single-stranded DNA (ssDNA) with free 3′ ends, facilitating strand exchange mediated by RecA protein.
- Its unwinding activity, coupled with other proteins, may provide a functional synergy necessary for effective recombination.
- RecBCD Enzyme:
- The RecBCD enzyme is a multifaceted complex that combines helicase, exonuclease, and ATPase activities, critical for homologous recombination.
- It unwinds DNA at a high rate, generating ssDNA substrates that enable RecA-mediated recombination, highlighting its essential role in DNA repair and genetic exchange.
- RuvAB Complex:
- This complex is involved in the processing of recombination intermediates, facilitating branch migration during homologous recombination.
- It acts after the formation of heteroduplex DNA, underscoring its role in the late stages of recombination.
- UvrAB Complex:
- Other Helicases:
- Helicase I:
- Encoded on the F plasmid, Helicase I is necessary for conjugative DNA transfer, featuring both nicking and unwinding activities that facilitate this process.
- Its unique dual function showcases the diverse roles helicases can play beyond DNA replication and repair.
- Helicase I:
Eukaryotic DNA Helicases
Eukaryotic DNA helicases play a crucial role in nucleic acid metabolism, primarily involved in essential processes such as DNA replication, repair, and recombination. While significant advancements have been made in understanding the function of these enzymes in simpler organisms like bacteria, much remains to be explored in the realm of eukaryotes. The intricacies of eukaryotic DNA helicases are particularly evident in the model organism Saccharomyces cerevisiae, or budding yeast, which serves as a valuable system for genetic and biochemical investigations.
- Understanding DNA Helicases: DNA helicases are enzymes responsible for unwinding the double-stranded DNA helix, allowing access for other proteins involved in replication and repair. Their activity is fundamental to ensuring the integrity of the genetic material during cellular processes.
- Challenges in Eukaryotic Research: Despite the importance of DNA helicases, research in eukaryotic systems has been hindered by difficulties in genetic analysis and the purification of sufficient quantities of the enzymes. These obstacles have limited the understanding of their specific roles in cellular processes.
- Characterization in Yeast: Research has identified at least 21 DNA helicases in eukaryotic organisms, with significant focus on S. cerevisiae. Genetic studies combined with biochemical techniques have allowed for the characterization of three notable helicases in yeast:
- Rad3: The RAD3 gene is critical in the excision repair pathway, particularly during the incision step. The Rad3 protein demonstrates 5′ to 3′ helicase activity and is crucial for unwinding damaged DNA strands. Mutations in RAD3 can lead to UV sensitivity, emphasizing its role in DNA repair.
- Pif1: The Pif1 protein is essential for mitochondrial DNA maintenance. Its helicase activity, also 5′ to 3′, is crucial for unwinding DNA, although challenges remain in its purification due to its mitochondrial localization. Pif1 function overlaps with the mitochondrial single-stranded binding protein Rim1, indicating a collaborative role in mitochondrial DNA stability.
- Srs2: Isolated as a suppressor of UV-sensitive mutants, the SRS2 gene product acts in error-prone repair pathways during mitotic growth. It exhibits 3′ to 5′ helicase activity, making it unique among yeast helicases. This suggests a specific role in facilitating DNA unwinding during repair processes.
- Diversity of Helicases: Additional helicases have been identified in yeast, often associated with replication machinery. For instance, a 60 kDa helicase remains associated with yeast replication factor C and unwinds DNA in a 5′ to 3′ direction. Moreover, two additional helicases capable of unwinding extensive duplex substrates have been identified, although their gene assignments remain unresolved.
- Insights from Mammalian Systems: In mammalian cells, several helicases have been characterized from sources such as calf thymus and HeLa cells. For example, four helicases (A, B, C, D) have been partially purified and characterized, demonstrating various unwinding polarities and substrate preferences. Notably, helicase E and DNA helicase a are distinct entities, exhibiting their own unwinding characteristics influenced by single-stranded binding proteins.
- Role in Excision Repair: Genetic and biochemical studies suggest that certain helicases, like the one encoded by the ERCC3 gene, are linked to both DNA repair and transcription initiation. This dual role highlights the complex interplay between DNA maintenance and gene expression in eukaryotic cells.
- Future Directions: To gain deeper insights into eukaryotic DNA helicases, further biochemical characterization and genetic studies are needed. The ability to manipulate model organisms like yeast allows researchers to dissect the genetics and biochemistry of helicases, ultimately leading to a better understanding of their roles in cellular processes.
Functions of DNA Helicases
The following are the key functions of DNA helicases:
- DNA Replication:
- Helicases play a central role in DNA replication by unwinding the dsDNA to provide single-stranded templates for replication. In collaboration with single-stranded binding proteins, primase, and DNA polymerase, helicases ensure the smooth progression of the replication fork. Enzymes like PcrA1, RepA, and DnaB are involved in this process.
- Okazaki Fragment Maturation:
- On the lagging strand of replicating DNA, helicases such as Dna2 and WRN are essential for the maturation of Okazaki fragments. These fragments are short DNA sequences synthesized discontinuously, and helicases help resolve the structures formed during their synthesis and ligation.
- DNA Repair:
- DNA helicases are involved in several DNA repair pathways, including nucleotide excision repair and homologous recombination. For example, XPB and XPD helicases assist in nucleotide excision repair by unwinding the DNA at the site of damage, facilitating the removal of incorrect or damaged nucleotides. Additionally, helicases such as BLM and WRN play roles in DNA damage sensing and signaling for repair activation.
- Transcription Regulation:
- Helicases are critical in transcription initiation and termination. The Rho helicase, for instance, unwinds specific regions of dsDNA to make the gene accessible for transcription by RNA polymerase. It also regulates transcription termination by removing RNA from the DNA template once the synthesis of messenger RNA (mRNA) is complete. Helicases like TFIIH also assist in activating transcription factors and ensuring proper initiation.
- Recombination:
- During homologous recombination, helicases such as RecBCD in E. coli are necessary for strand exchange and recombination completion. These enzymes help maintain genome stability by facilitating the correct exchange of genetic information between homologous chromosomes.
- Telomere Maintenance:
- Helicases such as BLM and Pif1 are crucial in preventing telomere shortening, a process that occurs during DNA replication. By maintaining telomere length, these enzymes help preserve chromosome integrity and prevent premature cellular aging.
- Structural Maintenance and DNA Stability:
- Some helicases, referred to as structural helicases, play a role in maintaining the stability and correct architecture of DNA. They identify abnormal DNA structures such as loops, secondary structures, or knots and signal for repair. This function is critical for avoiding mutations and maintaining genomic integrity.
- DNA Damage Recognition and Signaling:
- Certain helicases act as sensors for DNA damage. They recognize unusual DNA structures or lesions and trigger the appropriate repair pathways, ensuring the cell responds to DNA damage promptly to avoid mutations and genomic instability.
- Chromatid Remodeling:
- Helicases like Rad54 and ATRX participate in chromatid remodeling, a process essential for maintaining proper chromatin structure. This remodeling is necessary for gene expression regulation, DNA replication, and repair.
- Translation and Termination:
- Some helicases, like eIF4A and Rho, are involved in the unwinding of RNA secondary structures during translation. They help ensure proper translation initiation and termination, facilitating the synthesis of proteins from mRNA templates.
| Biological Function of Helicases | Helicases |
|---|---|
| Replicative Helicases | PcrA1, RepA, UvrD, Dda, HSV UL5, HSV UL9, DnaB, PriA, T7gp4A, T7gp4B, T4gp41, SV40, TAG, Polyoma TAG, BPV E1, MCM 4/6/7, Dna2, FFA-1, RecD, TraI, NS3, RecQL4 |
| Repair Helicases | UvrD, UvrAB, PcrA, Rad3, helicase E, XPD, XPB, Dna2, RecD2, BACH1, HDH II, RecQ, WRN, Rtel1, BLM, RuvB, Mph 1, CHD4 |
| Recombination Helicases | RecBCD, RecG, RecQ, RuvAB, PriA, UvrD, T4 UvsW, HDH II, HDH IV, WRN, Tra I, Rho, PDH65, BLM, Srs2, Sgs1, Rtel1 |
| Other Functions of Helicases | |
| Transcription | SWI2, SNF2, TFIIH, Rho, Factor 2, TRCF, RecQL5, ERCC6/RAD26 |
| Translation | HSV UL5, eIF4A, RHA, Ded1p, vasa |
| Chromatin Remodeling | Rad54, ATRX, BLM, CHD4 |
| Maintenance of Telomeres | Pif1, Dna2, Rtel 1, WRN, BLM, FANC |
| Okazaki Fragment Maturation | Dna2, Pif1, WRN |
Functions of Helicases
The following points provide a comprehensive overview of the diverse functions of helicases, emphasizing their critical roles in cellular metabolism.
- DNA Replication:
- Helicases are integral to nucleic acid replication, facilitating the unwinding of double-stranded DNA (dsDNA) to create ssDNA templates required for synthesis. This unwinding occurs during three primary phases: initiation, elongation, and termination.
- Various helicases, including PcrA, DnaB, and T7gp4, participate in these processes, collaborating with other proteins such as primases and polymerases to ensure efficient replication.
- DNA Repair:
- Helicases play a crucial role in DNA repair by unwinding damaged regions of DNA, which is a prerequisite for most repair mechanisms that rely on ssDNA.
- Notable repair helicases, such as UvrD, XPD, and WRN, are among the first responders to DNA lesions, ensuring that the repair machinery can access the damaged sites.
- Recombination:
- During homologous recombination, helicases are involved in the unwinding of DNA structures, which is essential for both initiation and branch migration. Key helicases like RecBCD and RuvAB facilitate the exchange and repair of genetic material.
- Transcription:
- Helicases also function during transcription, where they help to unwind dsDNA, allowing RNA polymerases to access the ssDNA template required for RNA synthesis.
- Specific helicases, such as TFIIH and SNF2, are crucial for various stages of transcription, including initiation and elongation. Additionally, they play roles in the maintenance and termination of transcription processes.
- Translation:
- In the realm of translation, helicases are involved in unwinding RNA structures to ensure proper ribosome function. For instance, helicases like eIF4A facilitate the unwinding of mRNA, allowing ribosomes to initiate protein synthesis.
- Chromatin Remodeling:
- Helicases contribute to ATP-dependent chromatin remodeling, which alters chromatin structure to regulate access to DNA. This function is essential for various nuclear processes, including transcription and DNA repair.
- Helicases such as Rad54 and ATRX are notable for their roles in facilitating the remodeling of chromatin in response to metabolic demands.
- Maintenance of Telomeres:
- Telomere maintenance is another critical function of certain helicases, including Pif1 and WRN. These helicases prevent the shortening of telomeres during DNA replication, thereby playing a role in cellular aging and stability.
- Okazaki Fragment Maturation:
- During DNA replication, helicases are essential for the maturation of Okazaki fragments, which are short segments synthesized on the lagging strand. Dna2 and Pif1 are examples of helicases that assist in this process by unwinding the fragments to facilitate their eventual ligation.
Examples of DNA Helicases
Below are examples of helicases involved in key aspects of nucleic acid metabolism.
- DNA Replication
- DnaB (E. coli): This hexameric helicase (SF4 family) operates with 5′ to 3′ directionality and forms a crucial part of the replisome during DNA replication. Its role includes unwinding DNA at the replication fork, enabling the synthesis of the leading strand in tandem with DNA polymerase.
- Mcm Complex (Eukaryotes and Archaea): The minichromosome maintenance (Mcm) complex is a heterohexamer in eukaryotes (Mcm2–Mcm7) and a homohexamer in archaea. It functions in the initiation and elongation phases of DNA replication, unwinding double-stranded DNA to facilitate the synthesis of single-stranded templates for DNA polymerase.
- DNA Repair
- UvrD (E. coli): This SF1 helicase functions in DNA repair, specifically mismatch repair (MMR). It unwinds the DNA around a nick created by the mismatch repair machinery, enabling the removal of erroneous DNA sequences by exonuclease digestion. UvrD is also involved in nucleotide excision repair (NER), where it unwinds the damaged DNA strand.
- XPD (Humans): A component of the TFIIH complex, XPD is involved in transcription-coupled NER. Mutations in XPD can lead to disorders such as xeroderma pigmentosum and trichothiodystrophy, highlighting its role in maintaining DNA integrity.
- DNA Recombination
- RecBCD (E. coli): This complex is essential for homologous recombination and consists of two helicases with opposite directionality (RecB with 3′ to 5′ and RecD with 5′ to 3′ directionality). RecBCD helps process DNA breaks and initiates recombination by unwinding DNA and facilitating strand exchange.
- RecQ Family (Humans): Members of the RecQ helicase family, such as BLM, WRN, and RECQ4, play significant roles in maintaining genome stability through recombination regulation. Mutations in these helicases are linked to genetic disorders, including Bloom syndrome (BLM mutation) and Werner syndrome (WRN mutation).
- Transcription
- Rho (E. coli): This hexameric SF5 helicase terminates transcription by binding to specific RNA regions rich in cytosine residues. Rho’s RNA-dependent ATPase activity enables it to translocate along the nascent transcript, facilitating transcription termination by dissociating the RNA-DNA hybrid.
- RNA Helicase A (Dhx9): Involved in transcription regulation, this helicase unwinds RNA structures during transcription, linking transcription to RNA processing steps.
- Ribosome Biogenesis
- DExD/H-box Proteins: Numerous DEAD-box helicases participate in ribosome biogenesis by unwinding RNA during the processing of pre-rRNA. These helicases facilitate interactions between snoRNAs and pre-rRNA, aiding in RNA modification and processing within ribosomes.
- RNA Splicing
- Prp Proteins (Yeast): Helicases such as Prp2 and Prp16 are involved in specific steps of RNA splicing, where they unwind RNA duplexes and drive conformational changes in the spliceosome, allowing for precise removal of introns from pre-mRNA.
- Translation
- eIF4A: This DEAD-box helicase is involved in the initiation of translation by unwinding secondary structures in the 5’ untranslated region (UTR) of mRNA, thereby facilitating ribosome binding and efficient protein synthesis.
- Ded1 (Yeast) and Vasa (Drosophila): These helicases play roles in translation by remodeling RNA secondary structures to promote translation initiation.
- RNA Degradation
- RhlB (E. coli): A DEAD-box helicase that forms part of the RNA degradosome complex, RhlB facilitates RNA degradation by unwinding RNA structures during exonucleolytic digestion.
- Mtr4 (Eukaryotes): This DExH-box helicase is crucial for nuclear RNA degradation, participating in the exosome-mediated degradation of RNA molecules.
- Mitochondrial Genome Maintenance
- Twinkle: A mitochondrial DNA helicase responsible for mtDNA replication. It unwinds mtDNA and plays a crucial role in maintaining mitochondrial genome integrity, particularly in preventing the depletion of mtDNA copy numbers.
- Pif1: This SF1 helicase is involved in mitochondrial DNA repair and recombination, as well as regulating telomere activity in nuclear DNA, ensuring stability across both mitochondrial and nuclear genomes.
Helicase Disorders and Diseases
Below is an organized summary of notable helicase disorders and their implications.
- ATRX Helicas Mutations:
- The ATRX gene encodes a helicase involved in chromatin remodeling, gene regulation, and DNA methylation.
- Located on the X chromosome, ATRX plays a critical role in embryonic development and prevention of apoptosis.
- Mutations primarily occur in the zinc finger and helicase domains, leading to ATR-X syndrome, which is characterized by:
- Microcephaly and skeletal abnormalities
- Mental retardation and genital abnormalities
- Seizures and limited language skills
- Alpha-thalassemia, a condition affecting hemoglobin production
- ATR-X symptoms suggest that ATRX mutations downregulate gene expression, particularly of the alpha-globin genes. The exact mechanisms that cause varied phenotypes among patients remain unclear.
- XPD Helicase Point Mutations:
- The XPD gene encodes a 5′-3′, ATP-dependent helicase and is a crucial component of the TFIIH complex, which is essential for nucleotide excision repair.
- Mutations in XPD have been associated with several disorders, including:
- Xeroderma pigmentosum (XP): Characterized by extreme sensitivity to UV light and a dramatically increased risk of skin cancer due to defective DNA repair mechanisms.
- Cockayne syndrome (CS): Involves developmental abnormalities and severe mental retardation from birth, along with sensitivity to UV light and premature aging.
- Trichothiodystrophy (TTD): This disorder presents with brittle hair and also features symptoms of UV sensitivity and premature aging.
- XPD mutations often disrupt protein interactions necessary for the stability of the TFIIH complex, leading to failures in DNA repair and transcription. In particular, mutations in XPD linked to Cockayne syndrome may cause rigidity in the protein, preventing necessary transitions from repair to transcription modes.
- RecQ Family Mutations:
- The RecQ helicase family consists of several members (e.g., BLM, WRN, RECQL4), all of which contribute to genome stability by regulating DNA replication and repair processes.
- Mutations in these helicases can lead to autosomal recessive disorders, including:
- Bloom syndrome (BS): Characterized by early onset of cancer and a predisposition to chromosomal instability, with a mean age of onset around 24 years. Cells exhibit a high frequency of sister chromatid exchanges and chromosomal damage.
- Werner syndrome (WS): Marked by premature aging, with symptoms including atherosclerosis, osteoporosis, and a high incidence of age-related diseases and cancers. Patients often experience significant genomic instability.
- Rothmund-Thomson syndrome (RTS): Characterized by skin and skeletal abnormalities, early aging signs, and increased cancer risk, including osteosarcomas. Cells show notable chromosomal rearrangements.
- RecQ helicases play crucial roles in ensuring proper DNA metabolism, and their deficiencies can lead to genomic instability and associated disorders.
- RecQ Helicases in Model Organisms:
- RecQ helicases, such as Sgs1 in yeast, serve as model systems for studying human helicase functions and associated disorders. Research on these helicases aids in understanding how mutations contribute to genomic instability and aging.
- Implications of Helicase Disorders:
- Helicases are vital for DNA metabolism. Disorders arising from mutations in helicase genes often lead to severe consequences, including cancer predisposition, developmental disorders, and premature aging.
- Research into the molecular mechanisms of helicase function can inform therapeutic approaches for these genetic disorders, highlighting the importance of understanding helicase activity in both normal physiology and disease contexts.
- Raney, K. D., Byrd, A. K., & Aarattuthodiyil, S. (2012). Structure and Mechanisms of SF1 DNA Helicases. DNA Helicases and DNA Motor Proteins, 17–46. doi:10.1007/978-1-4614-5037-5_2
- Burnham, D.R., Kose, H.B., Hoyle, R.B. et al. The mechanism of DNA unwinding by the eukaryotic replicative helicase. Nat Commun 10, 2159 (2019). https://doi.org/10.1038/s41467-019-09896-2
- Huttner, D., & Hickson, I. D. (2013). Helicases. Brenner’s Encyclopedia of Genetics, 406–408. doi:10.1016/b978-0-12-374984-0.00687-2
- Abdelhaleem M. Helicases: an overview. Methods Mol Biol. 2010;587:1-12. doi: 10.1007/978-1-60327-355-8_1. PMID: 20225138.
- Brosh RM Jr, Matson SW. History of DNA Helicases. Genes (Basel). 2020 Feb 27;11(3):255. doi: 10.3390/genes11030255. PMID: 32120966; PMCID: PMC7140857.
- Matson SW, Bean DW, George JW. DNA helicases: enzymes with essential roles in all aspects of DNA metabolism. Bioessays. 1994 Jan;16(1):13-22. doi: 10.1002/bies.950160103. PMID: 8141804.
- https://studymind.co.uk/notes/dna-replication/
- https://study.com/academy/lesson/dna-helicase-definition-role-function.html
- https://www.sciencedirect.com/topics/agricultural-and-biological-sciences/dna-helicase
- https://en.wikipedia.org/wiki/Helicase
- https://www.tebubio.com/blog/what-type-of-biological-molecule-is-a-dna-helicase/
- https://geneticeducation.co.in/helicase-structure-function-and-mechanism-of-dna-unwinding/
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.