In the world of genetics, there exists a remarkable technique that allows us to unlock the mysteries hidden within our DNA. DNA fingerprinting, also known as DNA profiling, is a laboratory genetic method that has revolutionized the way we identify individuals, establish relationships, and investigate crimes. By analyzing specific regions of our unique genetic code, this technique enables us to create a distinctive genetic “fingerprint” for each person or organism. As we delve deeper into the intricacies of DNA fingerprinting, we will explore its definition, process, applications, advantages, and disadvantages. Join us on this enlightening journey to enhance your genetic knowledge and academic understanding.
- Nearly every cell in the human body contains DNA.
- Approximately 99.9 percent of the DNA between two humans is identical on average.
- The remaining percentage is what makes us unique (barring identical twins, of course!).
- Although this may seem like a minor number, it indicates that there are approximately three million base pair differences between two individuals. These differences can be compared and used to differentiate you from another individual.
- Minisatellites are brief DNA sequences (10-60 base pairs in length) that exhibit greater variation between individuals than other regions of the genome. This variation manifests itself in the number of repeated units or’stutters’ in the minisatellite sequence.
- In 1980, the first minisatellite was discovered.
- Professor Sir Alec Jeffreys devised DNA fingerprinting in 1984 after realising that variations in human DNA could be detected using these minisatellites.VNTRs and STRs
- DNA fingerprinting is a technique that simultaneously detects a large number of minisatellites in the genome to generate an individual-specific pattern. This is an example of a DNA fingerprint.
- It is extremely unlikely that two individuals with identical DNA fingerprints are not identical twins.
- Your DNA fingerprint, similar to your actual fingerprint, is something you are born with and is unique to you.
What is DNA fingerprinting? – DNA fingerprinting definition
DNA fingerprinting is a technique used to identify an individual from a DNA sample by analysing the individual’s unique DNA patterns.
- DNA fingerprinting, also known as DNA typing, DNA profiling, genetic fingerprinting, genotyping, or identity testing, is a technique used in genetics to isolate and identify variable elements within the base-pair sequence of DNA (deoxyribonucleic acid).
- Alec Jeffreys, a British geneticist, devised the technique in 1984 after observing that certain sequences of highly variable DNA (known as minisatellites) that do not contribute to the functions of genes are repeated within genes.
- Each individual has a unique pattern of minisatellites, with the exception of multiple individuals from a single zygote, such as identical twins.
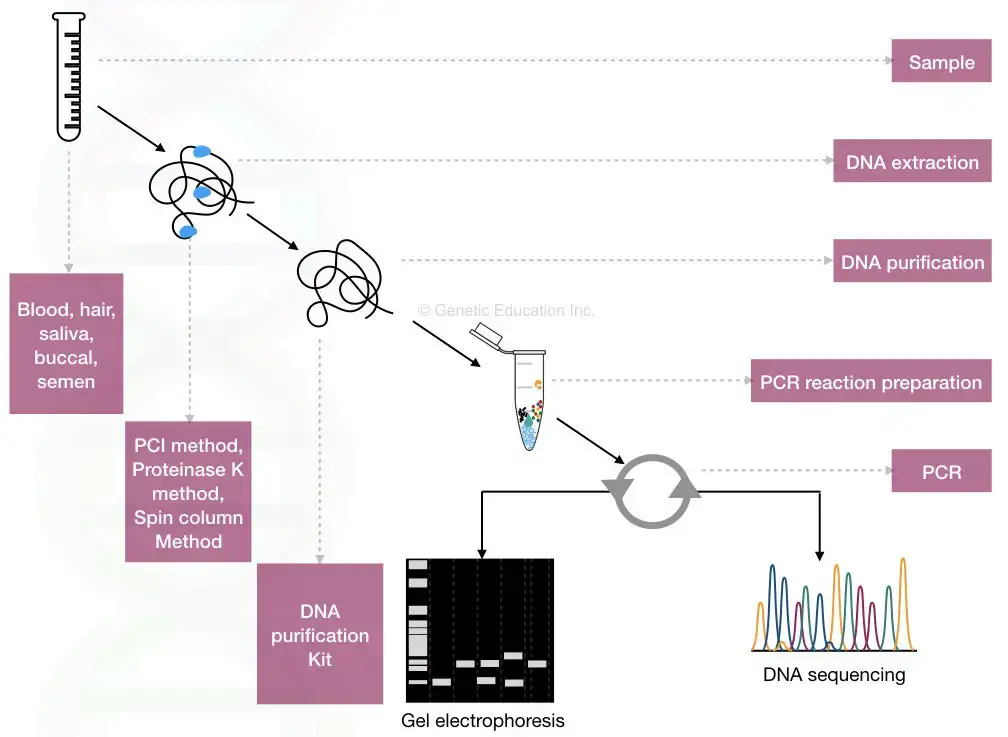
- The procedure for producing a DNA fingerprint begins with obtaining a sample of DNA-containing cells, such as skin, hair, or blood cells.
- The DNA is extracted and purified from the cells. In Jeffreys’s original method, which was based on restriction fragment length polymorphism (RFLP) technology, restriction enzymes were used to cut the DNA at specific locations along the strand.
- The enzymes generated fragments of differing lengths, which were sorted using electrophoresis: the shorter the fragment, the faster it moved towards the positive pole (anode). The double-stranded DNA fragments were then separated into single strands and transferred to a nylon sheet as part of a blotting technique.
- The fragments were subjected to autoradiography in which they were exposed to radioactive synthetic DNA probes that bound to the minisatellites. A piece of X-ray film was then exposed to the fragments, producing a dark mark wherever a radioactive probe had attached itself. The pattern of resulting markings could then be analysed.
- Microsatellites (or short tandem repeats, STRs) have shorter repeat units (typically 2 to 4 base pairs in length) than minisatellites (10 to over 100 base pairs in length). PCR multiplies the desired DNA fragment (e.g., a specific STR) in order to produce thousands of copies of it.
- It is an automated procedure that works with partially degraded DNA and requires only small quantities of DNA as starting material. Once a sufficient quantity of DNA has been produced using PCR, the exact sequence of nucleotide pairs within a DNA segment can be determined using one of several biomolecular sequencing techniques.
- Numerous new practical applications, such as identifying segments of genes that cause genetic diseases, mapping the human genome, engineering drought-resistant plants, and producing biological drugs derived from genetically modified organisms, have become possible as a result of the rapid acceleration of DNA sequencing made possible by the advent of automated equipment.
- An early application of DNA fingerprinting was in legal disputes, specifically to help solve crimes and establish paternity. Since its inception, DNA fingerprinting has led to the conviction of numerous criminals and the exoneration of numerous wrongfully convicted individuals.
- However, it is frequently difficult to make scientific identification coincide precisely with legal substantiation. Sometimes, even a single hint of the possibility of error is sufficient to convince a jury not to convict a suspect.
- Major sources of error include sample contamination, improper preparation procedures, and misinterpretation of results. In addition, RFLP requires significant quantities of high-quality DNA, limiting its use in forensics.
- Frequently, forensic DNA samples are degraded or collected postmortem, which makes them of inferior quality and susceptible to yielding less reliable results than samples obtained from a living individual. The development of PCR- and STR-based techniques alleviated a number of concerns regarding DNA fingerprinting, specifically the use of RFLP.
History of DNA fingerprinting
DNA fingerprinting, also known as DNA profiling or genetic fingerprinting, is a forensic technique used to identify individuals based on their unique DNA patterns. The history of DNA fingerprinting dates back to the 1980s and involves the contributions of several scientists.
- Discovery of DNA Structure: The foundation for DNA fingerprinting was laid with the discovery of the structure of DNA by James Watson and Francis Crick in 1953. Their work established that DNA consists of a double helix structure made up of nucleotide bases.
- DNA Variation: In the 1960s and 1970s, scientists discovered that while the majority of human DNA is the same in all individuals, certain regions exhibit variations in the number of repetitive DNA sequences. These regions, known as variable number tandem repeats (VNTRs), provided the basis for DNA fingerprinting.
- Alec Jeffreys and the First DNA Fingerprint: In 1984, British geneticist Sir Alec Jeffreys made a significant breakthrough in DNA fingerprinting. He discovered that certain sections of DNA contained highly variable and inherited VNTR sequences. These sequences, known as minisatellites or hypervariable regions, could be used to differentiate individuals.
- The Colin Pitchfork Case: The first high-profile application of DNA fingerprinting occurred in 1986 during the investigation of the double murder and rape of two teenage girls in the English village of Narborough. Geneticist Sir Alec Jeffreys was asked to analyze DNA samples from the crime scene and compare them to samples from potential suspects. The DNA evidence conclusively linked the suspect, Colin Pitchfork, to the crime, leading to his arrest and subsequent conviction in 1988. This case demonstrated the power of DNA fingerprinting as a forensic tool and highlighted its potential for solving crimes.
- Polymerase Chain Reaction (PCR): Another crucial development in DNA fingerprinting came with the invention of the polymerase chain reaction (PCR) by Kary Mullis in 1983. PCR allows for the amplification of small DNA samples, enabling the generation of sufficient DNA material for analysis.
- Short Tandem Repeats (STRs): In the late 1980s and early 1990s, researchers realized that using VNTRs in DNA fingerprinting had limitations due to the technical difficulties involved in analyzing them. This led to the discovery of a new type of genetic marker called short tandem repeats (STRs). STRs are shorter repetitive sequences of DNA that are more easily analyzed. They replaced VNTRs as the primary markers used in DNA fingerprinting.
- DNA Databases and Forensic Applications: In the 1990s, the establishment of DNA databases, such as the United Kingdom’s National DNA Database, provided a valuable resource for storing DNA profiles of individuals. These databases facilitate the comparison of DNA samples from crime scenes with known profiles, aiding in the identification of suspects and the resolution of criminal cases.
- Expansion of DNA Fingerprinting: Over time, DNA fingerprinting has found applications beyond forensic investigations. It is widely used in paternity testing, immigration cases, missing person identifications, and even studying genetic relationships among species in conservation efforts.
The development and refinement of DNA fingerprinting techniques have made it one of the most reliable and widely used forensic tools, revolutionizing the field of criminal justice and providing invaluable assistance in solving crimes and establishing personal identities.
Principle of DNA fingerprinting
DNA fingerprinting or DNA profiling identifies the combination of DNA sequences that frequently differ substantially between individuals.
- Short nucleotide repeats that vary in number between individuals and are inherited are the most important requirement for DNA fingerprinting. These are known as variable number tandem repeats or VNTRs.
- Except for identical twins (monozygotic twins), the DNA fingerprinting technique assumes that no two individuals have identical DNA sequences.
What is VNTRs and STRs?
Tandem repeats are sequences of DNA that are located one after another in the genome. They consist of repetitive units and can vary in length and number of repetitions from individual to individual. Two types of tandem repeats commonly used in DNA fingerprinting are Variable Number Tandem Repeats (VNTRs) and Short Tandem Repeats (STRs).
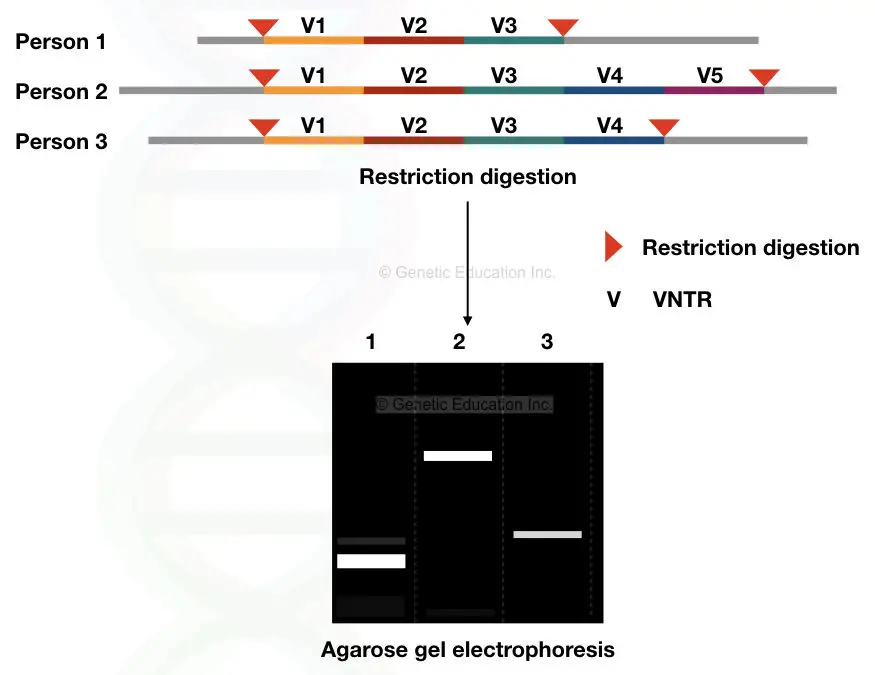
VNTRs are a type of minisatellite, ranging in size from 10 to 60 base pairs (bp), with 10 to 1000 repeats present in the genome. They produce a heterogeneous array of DNA fragments when analyzed. In the example provided, person “A” has 45 VNTRs with 20 bp repeats. The chances of finding another individual with the exact same number of repeats for this specific VNTR are extremely low, almost negligible, except in the case of monozygotic twins.
STRs, on the other hand, are a type of microsatellite. They are shorter in length, typically consisting of 1 to 6 bp repeats, and have 5 to 200 repeats in the genome. Compared to VNTRs, STRs produce a homogeneous array of DNA fragments during analysis.
To illustrate the concept further, let’s consider an example where different VNTRs (V1, V2, V3, V4, and V5) are digested with a single type of restriction endonuclease. The resulting fragments are then separated and visualized on an agarose gel using a technique called agarose gel electrophoresis. This technique allows for the detection of the size differences in the VNTR fragments and provides a DNA profile unique to each individual.
DNA fingerprinting using VNTRs and STRs has a high degree of accuracy and can distinguish between individuals with a probability of similarity as low as 1 in 10,000,000,000,000. This low probability reflects the extensive genetic variation in the human population, making DNA fingerprinting a powerful tool in forensic investigations, paternity testing, and other applications where individual identification is crucial.
| Aspect | VNTR (Variable Number Tandem Repeat) | STR (Short Tandem Repeat) |
|---|---|---|
| Number of Tandem Repeats | Variable | Short |
| Type of Repeat | Minisatellite | Microsatellite |
| Length of Repeat Units | 10 to 60 bp | 1 to 6 bp |
| Number of Repeats | 10 to 1000 | 5 to 200 |
| Array Produced | Heterogeneous | Homogeneous |
Methods of DNA fingerprinting
Three common methods are:
- RFLP based STR analysis
- PCR based analysis
- Real-time PCR analysis
1. RFLP based STR analysis for DNA fingerprinting
RFLP (Restriction Fragment Length Polymorphism) analysis was one of the initial methods used for DNA fingerprinting. Here’s the steps of RFLP-based STR analysis;
- Cleavage of DNA Fragments: In RFLP-based STR analysis, DNA is cleaved into fragments of various sizes using restriction endonucleases, which are enzymes that cut DNA at specific recognition sequences. The DNA fragments are then separated on an agarose gel based on their sizes.
- Southern Blotting: The separated DNA fragments on the gel are transferred to a nitrocellulose paper through a process called Southern blotting. This transfer allows for further analysis and manipulation of the DNA fragments.
- Chemical Denaturation: To immobilize the DNA on the nitrocellulose paper, chemical denaturation is performed. This process involves treating the DNA with chemicals to separate the DNA strands.
- Probe Hybridization: Radiolabeled DNA probes, which are complementary to specific target sequences, are used in probe hybridization. The probes bind to their complementary sequences in the denatured DNA on the nitrocellulose paper. This step helps in detecting specific genetic markers or sequences of interest.
- Incubation and Washing: The probes and the denatured DNA on the nitrocellulose paper are incubated together for several hours to allow for hybridization. Afterward, a washing step is performed to remove any unbound probes. This washing step is essential to eliminate non-specific bindings and ensure accurate results.
- X-ray Exposure and Results: The hybridized blot is then exposed to an X-ray film. Different DNA bands corresponding to the fragments with bound probes are observed on the X-ray film. These bands represent the presence of specific genetic markers or sequences, providing a unique pattern or “fingerprint” for each individual.
Limitations of RFLP-based STR Analysis: Although RFLP analysis was widely used for DNA fingerprinting, it had several limitations. The method was time-consuming, tedious, and expensive. Additionally, it required a larger quantity of DNA, making it less suitable for forensic samples, which often have limited or degraded DNA. These limitations led to the development and adoption of more efficient and sensitive techniques such as PCR-based STR analysis, which has largely replaced RFLP analysis in DNA fingerprinting applications.
2. PCR based analysis
PCR-based analysis, specifically PCR-based DNA fingerprinting, is a widely used and popular technique for genetic analysis. Here’s an overview of PCR-based analysis:
- Selection of STRs or VNTRs: The PCR-based DNA fingerprinting technique utilizes known STRs and VNTRs present in databases like the NCBI (National Center for Biotechnology Information). Researchers can select specific STR or VNTR sequences of interest for their analysis.
- Primer Design and PCR Amplification: Once the STR or VNTR sequence is chosen, specific primers are designed based on the sequence information. These primers are used to amplify the targeted DNA region during PCR (Polymerase Chain Reaction). PCR is a technique that enables the rapid amplification of specific DNA sequences.
- DNA Extraction and PCR Amplification: DNA extraction is performed to obtain the target DNA from the sample of interest. The extracted DNA is then subjected to PCR amplification using the designed primers. PCR exponentially amplifies the DNA region of interest, generating multiple copies for further analysis.
- Gel Electrophoresis Analysis: After PCR amplification, the amplified DNA fragments are analyzed using gel electrophoresis. Typically, agarose gel electrophoresis is used. The amplified DNA fragments are loaded onto the gel, and an electric current is applied. The DNA fragments migrate through the gel based on their size. Different-sized fragments result in distinct DNA bands on the gel.
- Differentiation of VNTRs and STRs: PCR-based gel electrophoresis is commonly used for VNTR analysis as VNTRs are larger fragments and can be separated effectively on the gel. VNTR analysis produces a distinctive band pattern on the gel. In contrast, STRs are shorter in length, making it more challenging to distinguish them using agarose gel electrophoresis. However, advancements in techniques such as capillary electrophoresis have improved the separation and analysis of both VNTRs and STRs.
- Capillary Electrophoresis: Capillary electrophoresis is a state-of-the-art method used in modern DNA analysis. It offers improved resolution and the ability to separate even small DNA fragments with high precision. Capillary electrophoresis is commonly employed for both VNTR and STR analysis due to its ability to discriminate fragments with a difference of a single base pair.
- Counting Repeats: While capillary gel electrophoresis is highly efficient in separating DNA fragments, it may not provide direct information on the exact number of repeats present in a DNA sequence. Additional techniques and analyses may be required to determine the precise number of repeats in VNTR or STR regions.
PCR-based analysis revolutionized DNA fingerprinting due to its simplicity, reliability, speed, and accuracy. It has become the preferred method for forensic DNA analysis, paternity testing, and various other applications requiring the identification and comparison of genetic profiles.
3. Real-time PCR analysis
Real-time PCR (Polymerase Chain Reaction) analysis, also known as quantitative PCR (qPCR), is a powerful technique that allows for the amplification of DNA while simultaneously quantifying the number of amplicons or DNA copies present in a sample. Here’s an overview of real-time PCR analysis:
- Amplification and Quantification: Real-time PCR not only amplifies the DNA but also enables the quantification of the amplified DNA in real-time. It can accurately determine the amount of template DNA present in a sample. This feature makes real-time PCR highly suitable for applications such as crime scene investigation, criminal verification, and other scenarios where precise DNA quantification is required.
- Fluoro-labeled Probes: Real-time PCR utilizes fluoro-labeled probes that specifically hybridize with the target DNA sequence of interest. These probes contain fluorescent labels that emit signals during the amplification process, allowing for the quantification of the target DNA. Unlike the probes used in traditional Southern blotting, the probes used in real-time PCR are not as long.
- Fluorescent Signal and Peaks: Based on the fluorescent signals emitted by the labeled probes, real-time PCR generates different peaks or curves that correspond to different DNA repeats or target sequences. The analysis of these peaks provides information about the quantity and characteristics of the DNA fragments present in the sample. Gel electrophoresis is not required in real-time PCR, making the technique faster and highly reliable.
- Calculation of Repeats: Real-time PCR enables the calculation of the number of DNA repeats present in different samples based on the fluorescent signals obtained. By comparing the peaks or curves generated, the number of repeats in the DNA can be determined. This information is valuable in various applications, such as genetic testing, forensic analysis, and paternity testing.
- Combination with Capillary Gel Electrophoresis: In forensic analysis, real-time PCR is often used in conjunction with capillary gel electrophoresis. This combination allows for both the quantification and separation of DNA repeats. Capillary gel electrophoresis provides high-resolution separation of DNA fragments based on their size, enhancing the accuracy and reliability of the analysis.
- DNA Sequencing: While real-time PCR is primarily focused on quantification, in other scientific applications, DNA sequencing is utilized to obtain detailed sequence information. DNA sequencing provides the complete nucleotide sequence of the target DNA, enabling researchers to study specific genetic variations and mutations.
Real-time PCR analysis is a rapid, accurate, reliable, and cost-effective method for DNA quantification and analysis. Its ability to simultaneously amplify and quantify DNA makes it an invaluable tool in various fields, including forensic science, medical research, and genetic diagnostics.
| Technique | Advantages | Disadvantages |
|---|---|---|
| RFLP | Accurate and cost-effective | Tedious and time-consuming process |
| Requires more DNA samples | ||
| PCR-agarose gel electrophoresis | Quick, easy to use, cheaper, accurate and reliable | Cannot determine the exact number of repeats |
| Cannot distinguish smaller DNA fragments | ||
| Does not provide sequence information | ||
| Rt-PCR | Fast, easy to use, cheap, accurate and measures repeats | Cannot distinguish smaller DNA fragments |
| Does not provide sequence information | ||
| Capillary electrophoresis | Can distinguish smaller DNA fragments with 1 bp change | Cannot determine the exact number of repeats |
| DNA sequencing | Provides sequence information | Time-consuming and costly |
DNA fingerprinting steps
- Collecting a biological specimen, such as blood, saliva, a buccal biopsy, sperm, or solid tissue.
- DNA isolation
- Choosing between RFLP, PCR amplification, real-time analysis, and capillary gel electrophoresis.
- Analysis and interpretation of results.
Process of DNA fingerprinting
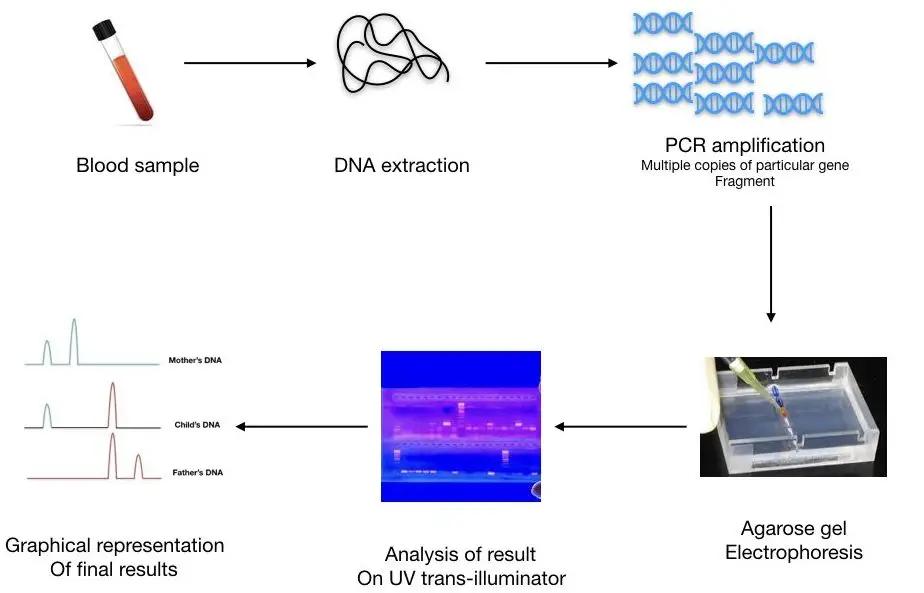
Step 1: Sample Collection: Various types of samples can be used for DNA collection, including buccal smears, saliva, blood, amniotic fluid, chorionic villi, skin, hair, body fluids, and other tissues. In criminal cases, buccal swabs are commonly used as they are non-invasive, although blood samples can also be used.
Step 2: DNA Extraction: DNA extraction is a crucial step to obtain DNA for analysis. Using a DNA extraction kit is recommended to ensure good quality and quantity of DNA. The extracted DNA should have sufficient purity and quantity for accurate results. Purification and quantification of DNA are performed using a DNA purification kit and UV-Visible spectrophotometer, respectively.
Step 3: Selecting a Technique: The choice of technique depends on the specific application. PCR-based gel electrophoresis is suitable for detecting maternal cell contamination, while capillary gel electrophoresis and real-time PCR are employed in forensic analysis. Each technique has its advantages and limitations, such as accuracy, ease of use, cost, ability to count repeats or distinguish smaller fragments, and the provision of sequence information.
Step 4: Results and Interpretation: The results and analysis process vary depending on the chosen technique. RFLP (Restriction Fragment Length Polymorphism) analysis involves observing DNA bands on autoradiography films. PCR-based gel electrophoresis visualizes DNA bands on a gel. Real-time PCR generates an amplification curve indicating the presence of different amplicons. DNA sequencing provides detailed sequence information of the target DNA. Capillary gel electrophoresis can separate even minute DNA bands and distinguish them.
| Technique | Results |
|---|---|
| RFLP | Autoradiography – DNA bands are observed on a film |
| PCR | Agarose gel electrophoresis – DNA bands are seen on a gel |
| rt-PCR | Amplification curve shows different amplicons in a sample |
| DNA sequencing | Provides sequence information of the target DNA |
| Capillary gel electrophoresis | Separates and distinguishes even minute DNA bands |
By comparing DNA profiles from different samples, information about variations and similarities between individuals can be obtained. Modern DNA fingerprinting processes are mostly automated, with computer analysis providing the final results.
DNA fingerprinting is a powerful tool used in various fields, including forensic science, paternity testing, and genetic research. It allows for the identification and differentiation of individuals based on their unique DNA profiles, providing valuable information for criminal investigations, family relationships, and genetic studies.
Applications of DNA Fingerprinting
The revolutionary technique of DNA fingerprinting finds application in various fields, ranging from forensic to medical science. The following are some popular applications of DNA fingerprinting:
- Revealing a person’s identity: DNA analysis provides a reliable method for determining a person’s biological identity. It is a highly accurate option available to establish individual identity.
- Identifying dead bodies: DNA fingerprinting can even be used to identify deceased individuals in cases of mass disasters. It helps identify badly damaged bodies by comparing available data with DNA profiles.
- Identification of blood relatives: DNA fingerprinting assists in establishing blood relationships between unrelated individuals. By comparing DNA samples from potential relatives, the technique determines whether they share a biological connection.
- Crime scene investigation and criminal verification: DNA fingerprinting plays a crucial role in forensic investigations and criminal verification. Samples collected from crime scenes, such as saliva, blood, or hair follicles, undergo DNA analysis to establish a link between the suspect and the crime.
- Organ transplantation studies: DNA profiling is utilized in organ transplantation to assess graft acceptance and rejection. The HLA typing technique is employed to match the Human Leukocyte Antigen (HLA) types of the donor and recipient, ensuring compatibility for successful organ transplantation.
- Maternal cell contamination detection: DNA fingerprinting is used to detect maternal cell contamination (MCC) in prenatal studies and diagnosis. Maternal cells present in chorionic villi samples or amniotic fluid can lead to false-positive results. DNA profiling helps identify and distinguish maternal cells, ensuring accurate genetic testing during pregnancy.
- Studying genetic diseases: DNA fingerprinting finds extensive applications in medical science, particularly in studying genetic diseases. Scientists use modified PCR techniques to analyze mutations, allele variations, and establish phylogenetic relationships among different organisms.
These applications demonstrate the versatility and importance of DNA fingerprinting across various fields, aiding in investigations, personal identification, medical research, and ensuring the accuracy of genetic testing.
How are Restriction enzymes used in DNA Fingerprinting?
- Restriction enzymes play a crucial role in DNA fingerprinting by aiding in the analysis of short tandem repeats (STRs). STRs are non-coding DNA sequences that contain repetitive elements located in the centromeric regions of chromosomes.
- To analyze STRs, DNA samples are first extracted from individuals of interest. These DNA samples contain the STR regions. Restriction enzymes, also known as restriction endonucleases, are then used to cleave the DNA at specific recognition sites within the STR regions.
- The restriction enzymes recognize specific DNA sequences and cut the DNA at or near these sites. Different restriction enzymes have different recognition sequences, allowing for targeted cutting of the DNA. The cleavage by restriction enzymes generates DNA fragments of different lengths, with the size variations occurring due to the different numbers of repeated sequences within the STR regions.
- After digestion with restriction enzymes, the DNA fragments are separated using a technique called gel electrophoresis. Gel electrophoresis involves placing the DNA fragments in a gel matrix and applying an electric field, causing the fragments to migrate through the gel based on their size. The smaller fragments move faster and travel farther, while the larger fragments move more slowly and remain closer to the starting point.
- The resulting pattern of DNA fragments on the gel represents the unique DNA profile of an individual. By comparing the DNA profiles of different individuals, it is possible to determine the similarities or differences in their STR regions. This information can be used in forensic investigations and paternity testing to establish identity or determine biological relationships.
- In summary, restriction enzymes are used in DNA fingerprinting to cleave the DNA at specific sites within the STR regions, allowing for the analysis and comparison of DNA profiles. The variations in the number of repeated sequences in STRs contribute to the uniqueness of each individual’s DNA profile.
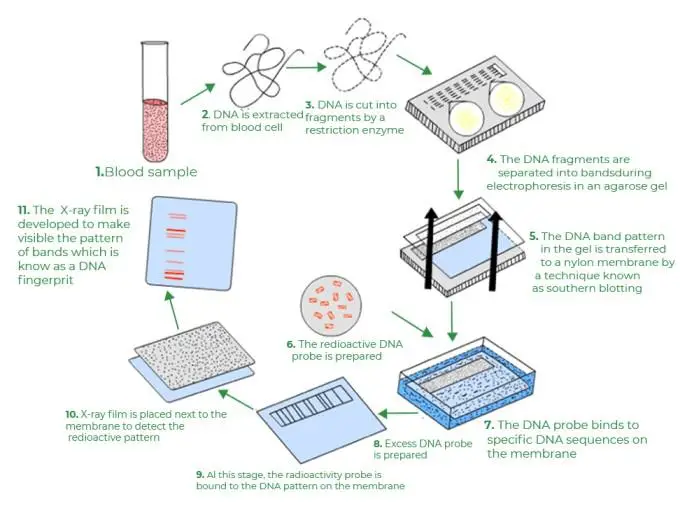
Simplified Process of DNA Fingerprinting
- Step 1: Isolation of DNA The cell’s DNA is extracted and purified through chemical processing and centrifugation.
- Step 2: Amplification Using polymerase chain reaction (PCR), numerous copies of the extracted DNA are created.
- Step 3: Digestion of DNA by Restriction Endonuclease Enzyme. The restriction endonuclease enzyme disassembles DNA into smaller fragments. These enzymes cleave the DNA at specific locations, resulting in fragments of varying lengths.
- Step 4: DNA Fragment Separation. Electrophoresis is then used to separate the DNA fragments according to their size. Electrophoresis is a technique used in the presence of an electric field to separate charged molecules. Electrophoresis involves placing DNA fragments on a transparent gel bed on a plate and administering an electric current to the gel. DNA fragments gravitate toward the positive electrode due to their distinct negative charges. Ultimately, the fragments and gel are distributed to numerous locations according to their sizes.
- Step 5 Introduce the agents that separate the DNA fragments into single strands.
- Step 6: Transferring the DNA fragments from the gel to synthetic membranes such as nylon or nitrocellulose (blotting). The DNA fragments are then transferred from the gel to a nylon membrane or nitrocellulose using the Southern blotting technique. This technique involves coating the gel with a nylon membrane that attracts DNA fragments, similar to how blotting paper dries damp ink.
- Step 7: Radiolabeled Probe Hybridizations. In this step, the radioactive isotope is hybridized with the DNA fragments so that their positions can be seen on an X-ray image. To achieve this, a nylon membrane is added to a bath containing probes (probes are short segments of single standard complementary DNA tagged with radioactivity that bind to a specific chain of DNA VNTR sequences based on the base-pairing rule).
- Step 8- detection of hybridized DNA fragments. The membrane is exposed to the X-ray film to produce an autoradiograph, which, when developed, reveals a distinct pattern of dark and brilliant bands that correspond to the DNA structure. On the X-ray film, the dark bands correspond to the DNA signatures.
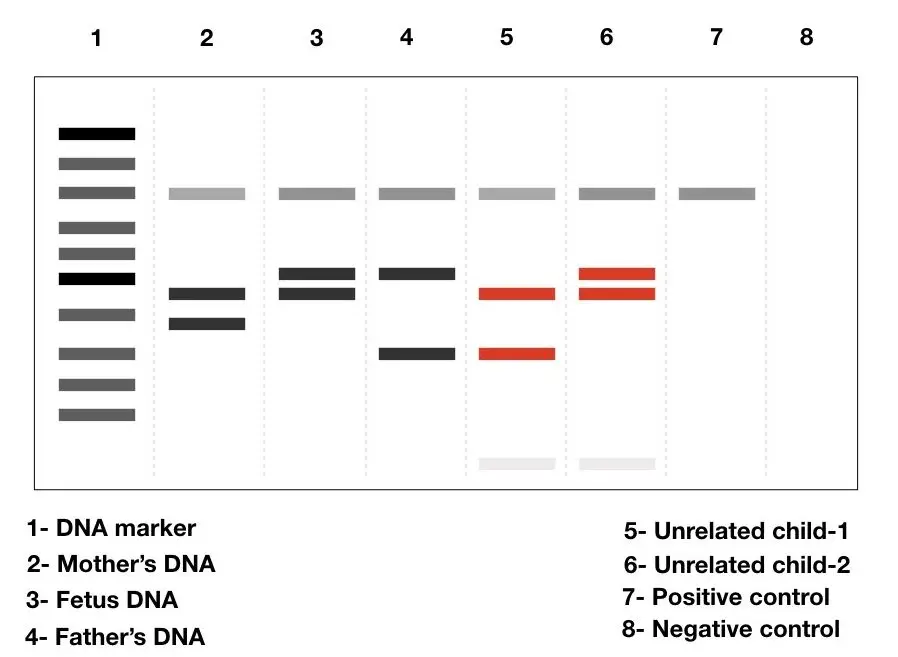
How we can create a DNA profile using STR
To create a DNA profile using Short Tandem Repeats (STRs), the following steps are typically followed:
Step 1: Obtain a sample of DNA A DNA sample can be collected from various sources such as blood, saliva, semen, hair follicles, or body tissues. Crime scene investigators collect DNA samples from crime scenes, while mouth swabs are commonly used to collect DNA directly from individuals by collecting cheek cells.
Step 2: Extract the DNA The DNA is extracted from the collected sample. This involves breaking open the cells to release the DNA and separating it from other cellular components using chemicals. The extracted DNA is purified to remove impurities and contaminants.
Step 3: Amplify the DNA using PCR Since forensic investigations often have limited amounts of DNA available, the DNA is amplified using a technique called Polymerase Chain Reaction (PCR). PCR allows for the selective amplification of specific DNA regions, including the STR regions. This process generates multiple copies of the DNA for further analysis.
Step 4: Analyze the size of the STRs The amplified DNA fragments containing the STR regions are then analyzed to determine the size of the STRs. This is typically done using a genetic analyzer, which detects the fluorescent dyes attached to the STRs. The DNA fragments are separated by size through a process called gel electrophoresis. The genetic analyzer reads the sizes of the STRs and generates a data profile.
Step 5: Compare and match the STRs The obtained DNA profile, consisting of the sizes of the STRs at different genetic loci, is compared to other DNA profiles. Forensic scientists typically analyze STRs at 10 or more genetic loci. If the STR sizes in the profiles match, it suggests that the samples originated from the same individual, providing strong evidence of a match.
By comparing STR profiles from different samples, forensic scientists can establish links between individuals, identify suspects in criminal investigations, determine biological relationships (such as paternity testing), and exclude individuals from consideration as the source of a DNA sample.
Overall, the use of STRs and the analysis of their sizes allow for the creation of a unique DNA profile that can be used for identification and forensic purposes.
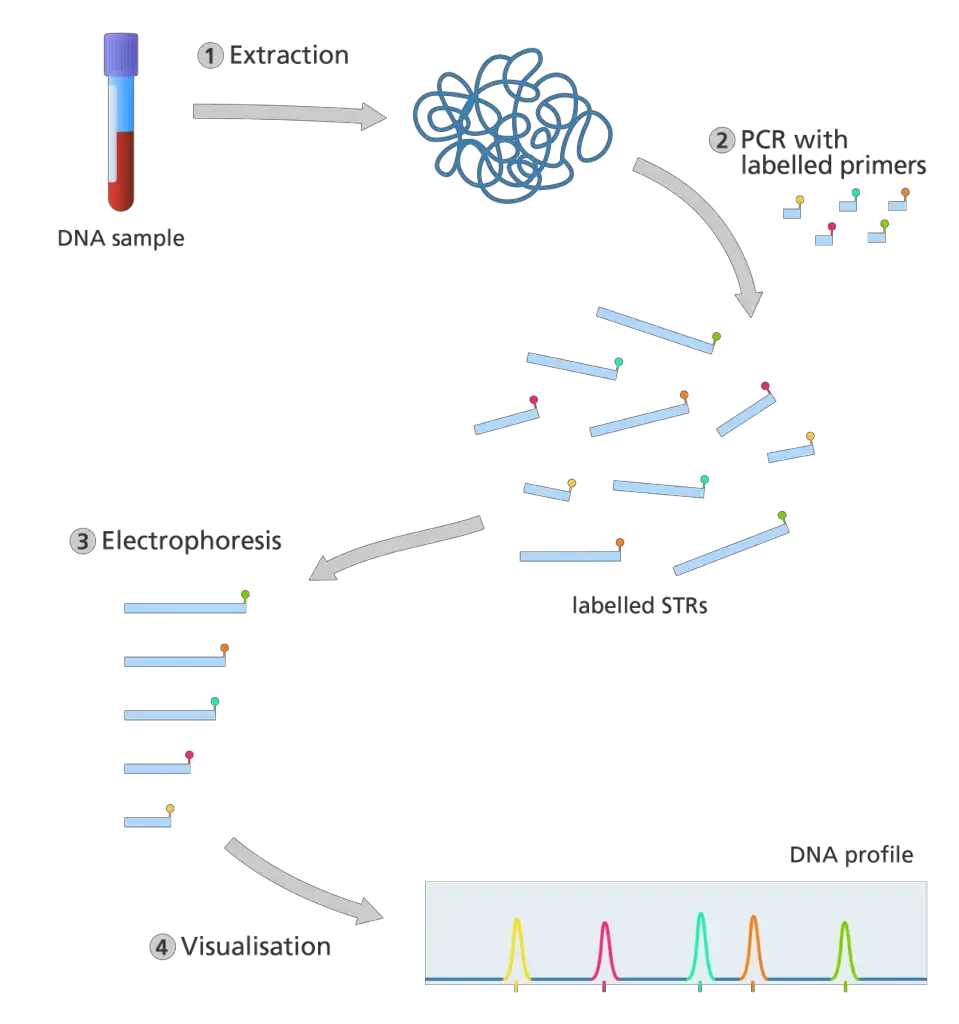
How is a DNA profile produced today?
- The extraction of DNA from a biological sample. STR analysis is extremely sensitive, so only a small amount of a person’s DNA is required to generate an accurate result. Thus, DNA can be extracted from a wider variety of biological samples, such as blood, saliva, and hair.
- DNA profiling, unlike the original DNA fingerprinting procedure, does not employ restriction enzymes to cut the DNA. In place of this, the polymerase chain reaction (PCR) is utilized to generate multiple copies of specific STR sequences.
- PCR is an automated method for producing multiple copies of a specific DNA sequence. It only requires small amounts of DNA to begin with and can create copies from partially degraded DNA samples.
- In polymerase chain reaction (PCR), primers bond to complementary sequences of the target DNA and serve as the starting point for copying the target DNA.
- In STR analysis, the PCR primers are designed to adhere to either end of the target STR sequence.
- The primers for each STR are fluorescently tagged with a specific color. This facilitates the identification and recording of STR sequences following PCR.
- Once sufficient copies of the sequence have been generated by PCR, electrophoresis is used to separate the fragments by size.
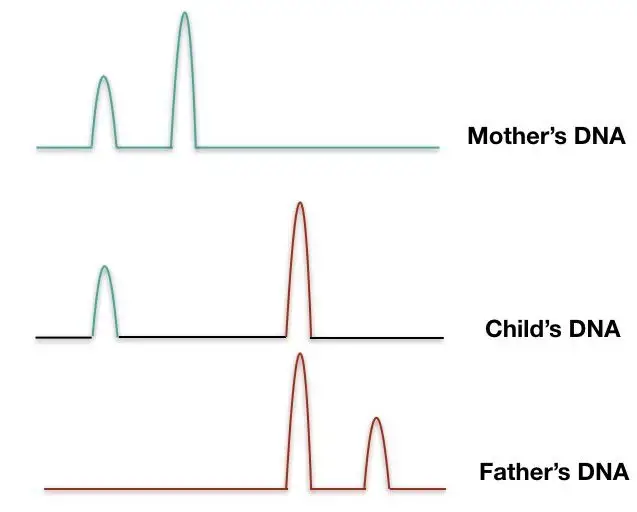
- Each fragment is passed by a laser, which causes fragments with fluorescent identifiers to emit a distinct color of light. The output is presented as a series of colored peaks (as depicted in the image below) that emphasize the color and length of each STR sequence.
How are DNA profiles stored?
- In 1995, the United Kingdom was the first nation to establish a national database of DNA profiles.
- The National DNA Database of the United Kingdom contains the DNA profiles of a select number of British citizens, the majority of whom are linked to severe crimes.
- The Protection of Freedom Act of 2013 mandated the removal of 1,766,000 innocent adult and juvenile DNA profiles from the UK National DNA Database.
- The majority of nations now maintain national DNA databases.
DNA fingerprinting pros and cons
Advanatages of DNA Fingerprinting/Pros of DNA Fingerprinting
DNA fingerprinting, also known as DNA profiling or genetic fingerprinting, offers several advantages in various fields. Some of the key advantages of DNA fingerprinting include:
- High Discriminatory Power: DNA fingerprinting is highly discriminatory and can distinguish between individuals with a high degree of accuracy. The use of specific genetic markers, such as short tandem repeats (STRs), allows for precise identification of unique DNA profiles, even among closely related individuals.
- Individual Identification: DNA fingerprinting provides a reliable method for individual identification. Each person has a unique DNA profile, except for identical twins who have identical DNA. This makes DNA fingerprinting highly valuable in forensic investigations, where it can be used to match crime scene evidence to suspects or establish the identity of unknown individuals.
- Paternity and Kinship Testing: DNA fingerprinting is extensively used in paternity and kinship testing to determine biological relationships between individuals. By comparing the DNA profiles of potential parents and offspring, it is possible to establish or exclude biological parentage with a high degree of accuracy.
- Forensic Investigations: DNA fingerprinting has revolutionized forensic investigations by enabling the analysis of DNA evidence recovered from crime scenes. It can link suspects to crimes, exonerate innocent individuals, and provide valuable evidence in court proceedings. DNA profiling has significantly enhanced the accuracy and reliability of criminal investigations.
- Missing Persons and Disaster Victim Identification: In cases of missing persons or mass disasters, DNA fingerprinting plays a crucial role in identification efforts. DNA samples from family members can be compared with recovered remains to establish positive identifications, providing closure to families and aiding in the identification of victims.
- Historical and Anthropological Studies: DNA fingerprinting has contributed to our understanding of human migration patterns, population genetics, and evolutionary history. By analyzing DNA profiles from different populations, scientists can infer relationships, track genetic variations, and unravel human ancestry.
- Wildlife Conservation and Forensics: DNA fingerprinting is valuable in wildlife conservation efforts. It can help identify and track endangered species, study population dynamics, investigate illegal wildlife trade, and provide evidence in wildlife-related crimes.
- Medical and Genetic Research: DNA fingerprinting plays a crucial role in medical and genetic research. It aids in the identification of disease-causing genes, studying genetic disorders, and assessing genetic predisposition to certain diseases. It is also used in pharmacogenomics, personalized medicine, and clinical trials.
Overall, DNA fingerprinting offers a highly accurate and reliable method for individual identification, kinship testing, and forensic investigations. Its wide range of applications makes it an indispensable tool in various fields, contributing to advancements in science, justice, and human understanding.
Limitations of DNA Fingerprinting/ Cons of dna fingerprinting
While DNA fingerprinting is a powerful and widely used technique, it also has certain limitations. Some of the limitations of DNA fingerprinting include:
- Contamination: DNA samples can easily become contaminated with external DNA, which can lead to inaccurate results. Even small amounts of extraneous DNA can interfere with the analysis and potentially produce false profiles.
- Degradation: DNA samples can degrade over time, especially in poorly preserved or older samples. Degraded DNA can result in incomplete or unreliable profiles, making it challenging to obtain conclusive results.
- Sample Size: DNA fingerprinting requires a sufficient quantity and quality of DNA for analysis. In cases where the DNA sample is limited or degraded, it may be challenging to generate a reliable profile. Low-quality samples may lead to inconclusive or ambiguous results.
- Mutations and Allelic Variants: Mutations and allelic variations can occur in the DNA sequences targeted for analysis, leading to variations in the results. These variations can affect the accuracy and reliability of DNA profiles, particularly in cases where the mutations or variants are present in the target loci.
- Population Genetics: DNA fingerprinting relies on the use of specific genetic markers, such as STRs, which can vary in frequency among different populations. This variation can affect the statistical interpretation of DNA profiles, particularly when comparing profiles between individuals from diverse ethnic backgrounds.
- Familial Relationships: DNA fingerprinting can determine biological relationships between individuals, such as parentage or sibling relationships. However, it may not provide definitive answers in complex cases involving distant relatives or situations where the genetic relationships are not straightforward.
- False Positives and False Negatives: While DNA fingerprinting is highly accurate, there is still a possibility of false-positive or false-negative results. Errors in sample handling, laboratory procedures, or interpretation can contribute to these errors, highlighting the need for rigorous quality control measures.
It is important to consider these limitations and potential sources of error when interpreting and relying on DNA fingerprinting results. The expertise of forensic scientists, adherence to strict protocols, and quality assurance procedures are crucial to minimizing these limitations and ensuring accurate and reliable DNA profiling.
How does dna fingerprinting work?
DNA fingerprinting, also known as DNA profiling or DNA typing, is a forensic technique used to analyze and compare DNA samples to identify individuals or determine their biological relationships. The process involves several key steps:
- Sample Collection: DNA samples can be collected from various sources, such as blood, saliva, hair follicles, semen, or body tissues. The samples are collected from crime scenes, suspects, victims, or individuals involved in paternity/maternity testing.
- DNA Extraction: The collected samples undergo DNA extraction to isolate the DNA molecules from other cellular components. This step involves breaking open the cells and releasing the DNA.
- Polymerase Chain Reaction (PCR): PCR is a technique used to amplify specific regions of the DNA sample. In DNA fingerprinting, Short Tandem Repeats (STRs) are targeted. STRs are regions of DNA with repeated sequences of 2 to 6 base pairs. PCR allows for the replication of these STR regions, generating a large quantity of DNA for analysis.
- Gel Electrophoresis: The amplified DNA fragments are separated and analyzed using gel electrophoresis. The DNA fragments are loaded onto a gel matrix and subjected to an electric field. Since DNA is negatively charged, it migrates towards the positive electrode. The gel acts as a sieve, separating the DNA fragments based on their size.
- DNA Visualization: After electrophoresis, the DNA fragments are visualized using a stain or fluorescent dye. This step allows the detection of the DNA bands or peaks, which represent the different alleles at specific STR loci.
- DNA Analysis: The DNA fingerprinting analysis involves comparing the patterns of DNA bands or peaks from different samples. The number and size of the bands at each STR locus are recorded, creating a unique DNA profile for each individual. The profiles are typically presented as a series of numbers representing the alleles at specific STR loci.
- Data Interpretation: The DNA profiles are analyzed to determine relationships or identify individuals. In forensic investigations, the DNA profile from a crime scene sample can be compared to known reference samples from suspects to establish a match or exclusion. In paternity/maternity testing, the profiles of the child and alleged parents are compared to determine biological relationships.
The uniqueness of DNA sequences and the high variability of STR regions make DNA fingerprinting a powerful tool for individual identification and relationship determination. The process has revolutionized forensic science, providing reliable and accurate evidence in criminal investigations, missing person cases, and paternity/maternity disputes.
How does restriction enzyme relate to dna fingerprinting?
Restriction enzymes play a crucial role in DNA fingerprinting. These enzymes, also known as restriction endonucleases, are proteins that can recognize specific DNA sequences and cut the DNA at or near those sequences. The use of restriction enzymes is an essential step in DNA fingerprinting. Here’s how they relate to the process:
- DNA Fragmentation: In DNA fingerprinting, the DNA sample is first fragmented to create smaller DNA fragments. Restriction enzymes are used to selectively cut the DNA at specific recognition sites within the genome. These recognition sites are typically short sequences of 4 to 8 base pairs that are palindromic, meaning they read the same in both directions (e.g., GAATTC). When the restriction enzyme encounters its target sequence, it cuts the DNA, resulting in the fragmentation of the DNA molecule at specific sites.
- Creating DNA Profiles: The fragmented DNA generated by the restriction enzyme digestion is then subjected to gel electrophoresis. Gel electrophoresis separates the DNA fragments based on their size. The DNA fragments are loaded onto a gel matrix, and an electric current is applied. Since DNA is negatively charged, it migrates through the gel towards the positive electrode. The smaller DNA fragments move faster through the gel, while the larger fragments move more slowly. This separation of DNA fragments creates a pattern of bands on the gel, known as a DNA profile.
- Comparison and Identification: The DNA profiles obtained from different individuals can be compared to determine similarities and differences. The presence or absence of specific DNA bands at certain locations on the gel can be used to identify individuals or establish relationships. If two individuals have identical or highly similar DNA profiles, it indicates a potential match or a close biological relationship.
- Unique Patterns: The use of restriction enzymes in DNA fingerprinting helps create unique DNA profiles for each individual. The locations of the recognition sites and the resulting fragment sizes vary among individuals due to genetic variations. This variation leads to distinct patterns of DNA bands on the gel, which are unique to each individual (except for identical twins). Therefore, DNA fingerprinting can provide a reliable method for individual identification.
In summary, restriction enzymes are employed in DNA fingerprinting to fragment the DNA, which is then separated by gel electrophoresis to generate DNA profiles. These profiles, with their unique banding patterns, can be compared to identify individuals or determine relationships in forensic investigations, paternity/maternity testing, and other applications of DNA analysis.
How does dna fingerprinting distinguish one individual from another?
DNA fingerprinting distinguishes one individual from another by analyzing specific regions of the DNA that exhibit variation between individuals. These regions are known as polymorphic regions, and they contain repetitive DNA sequences, such as short tandem repeats (STRs) or variable number tandem repeats (VNTRs).
The process of DNA fingerprinting involves several steps:
- DNA Sample Collection: A sample containing DNA is collected from the individual, typically through sources like blood, saliva, or hair follicles.
- DNA Extraction: The DNA is extracted from the sample, isolating it from other cellular components.
- Polymerase Chain Reaction (PCR): Specific regions of the DNA, typically STRs, are amplified using a technique called PCR. PCR utilizes primers that flank the target regions and replicate them millions of times, generating numerous copies of the DNA fragments.
- Gel Electrophoresis: The amplified DNA fragments are separated by size using gel electrophoresis. The DNA fragments are loaded onto a gel and subjected to an electric current. Smaller fragments migrate faster through the gel, while larger fragments move slower, creating distinct bands.
- Visualization and Analysis: The separated DNA fragments are visualized using methods like staining or fluorescent labeling. The resulting pattern of bands on the gel represents the individual’s DNA profile or fingerprint.
Each individual has a unique combination of DNA fragment sizes at specific regions. By comparing the DNA profiles of different individuals, scientists can determine whether they match or differ. If the patterns of DNA bands at multiple loci match between two samples, it indicates that the DNA came from the same individual. Conversely, differences in the DNA band patterns indicate that the samples came from different individuals.
The probability of two unrelated individuals having the same DNA profile is extremely low, making DNA fingerprinting a powerful tool for identification purposes. The analysis typically examines multiple loci to enhance the accuracy and reliability of distinguishing individuals.
Overall, DNA fingerprinting relies on the variability of specific DNA regions among individuals to establish unique patterns that distinguish one individual from another.
DNA fingerprinting worksheet
dna fingerprinting worksheet answer key
FAQ
What 2 famous murder investigations was helped by dna fingerprinting?
Two famous murder investigations that were significantly aided by DNA fingerprinting are the following:
O.J. Simpson Trial (1995): The trial of former American football player O.J. Simpson for the murders of his ex-wife Nicole Brown Simpson and her friend Ronald Goldman is notable for its use of DNA fingerprinting evidence. DNA analysis conducted on blood samples found at the crime scene, including bloodstains on clothing and the infamous bloody glove, provided crucial evidence linking O.J. Simpson to the murders. The DNA profiles obtained from the samples matched O.J. Simpson’s DNA, which played a pivotal role in the prosecution’s case and ultimately contributed to his acquittal.
Colin Pitchfork Case (1988): The Colin Pitchfork case, which occurred in the United Kingdom, marked the first time DNA fingerprinting was used to solve a murder investigation. In 1986 and 1987, two young girls were raped and murdered in Leicestershire. DNA evidence recovered from the crime scenes was compared with DNA samples obtained from suspects. The groundbreaking DNA fingerprinting technique, developed by geneticist Sir Alec Jeffreys, played a crucial role in identifying the perpetrator. Colin Pitchfork became the first person to be convicted of murder based on DNA fingerprinting evidence in 1988, leading to a significant advancement in forensic science.
These cases highlight the pivotal role that DNA fingerprinting played in providing conclusive evidence, identifying the perpetrators, and influencing the outcomes of these high-profile murder investigations. DNA fingerprinting has since become an essential tool in forensic investigations worldwide, aiding in solving numerous crimes and delivering justice.
When was dna fingerprinting invented?
DNA fingerprinting, also known as DNA profiling or DNA typing, was invented in 1984 by British geneticist Sir Alec Jeffreys. While conducting research at the University of Leicester, Jeffreys discovered a unique pattern of DNA fragments that could be used to distinguish individuals. His breakthrough came when he applied a technique called DNA hybridization to analyze minisatellite DNA sequences, which are highly variable regions of the genome. This discovery paved the way for the development of DNA fingerprinting as a powerful tool in forensic science, paternity testing, and other fields. Sir Alec Jeffreys’ pioneering work in DNA fingerprinting revolutionized the field of genetics and has had a profound impact on various areas of science and law enforcement.
In dna fingerprinting what do restriction enzymes do?
In DNA fingerprinting, restriction enzymes play a crucial role in the process. Restriction enzymes, also known as restriction endonucleases, are proteins that can recognize specific DNA sequences and cut the DNA at those sites. These enzymes are naturally found in bacteria as a defense mechanism against viral DNA.
In the context of DNA fingerprinting, restriction enzymes are used to create DNA fragments with specific patterns. The DNA sample, which contains the target regions of interest, is treated with a specific restriction enzyme that recognizes and cuts at specific recognition sites within the DNA sequence. The recognition sites are typically short sequences, often palindromic, meaning they read the same forward and backward.
When the restriction enzyme cuts the DNA at these recognition sites, it generates fragments of different lengths. The distribution of these fragment lengths is unique to each individual, as the locations of the recognition sites and the lengths of the DNA fragments vary between individuals due to genetic differences.
After the DNA is cut by the restriction enzyme, the fragments are separated using a technique called gel electrophoresis. The DNA fragments are loaded onto a gel and subjected to an electric current, causing the fragments to move through the gel based on their size. This separation allows for the visualization of the DNA fragments as distinct bands on the gel.
By comparing the patterns of DNA bands obtained from different individuals, scientists can create a DNA fingerprint or profile. The DNA fingerprint represents the unique arrangement of DNA fragments for an individual, which can be used for identification purposes, such as in forensic investigations, paternity testing, and genetic research.
Which type of dna is used during dna fingerprinting?
During DNA fingerprinting, a specific type of DNA called nuclear DNA is used. Nuclear DNA is located within the nucleus of cells and contains the majority of an individual’s genetic information. It is inherited from both parents and is present in almost all cells of the body.
Nuclear DNA is preferred for DNA fingerprinting because it carries the unique genetic code that distinguishes one individual from another. Within the nuclear DNA, specific regions known as short tandem repeats (STRs) or variable number tandem repeats (VNTRs) are targeted for analysis. These regions consist of repeated sequences of nucleotides, and the number of repeats varies between individuals, making them useful for identifying genetic differences.
The DNA samples collected for DNA fingerprinting can be obtained from various sources, such as blood, saliva, hair follicles, or tissue samples. The DNA is extracted from these samples, and specific regions containing STRs or VNTRs are amplified using a technique called polymerase chain reaction (PCR). The amplified DNA fragments are then separated and visualized using gel electrophoresis or other analytical methods, allowing the comparison of DNA profiles between individuals.
By analyzing the variation in the number and size of the repeated sequences at specific loci, DNA fingerprinting can generate unique patterns or profiles for each individual, which can be used for identification, forensic investigations, and relationship testing.
DNA fingerprinting would most likely be used to?
DNA fingerprinting is most commonly used for the following purposes:
Forensic Investigations: DNA fingerprinting plays a crucial role in forensic science to identify suspects and link them to crime scenes. DNA samples collected from crime scenes, such as blood, semen, or hair, can be compared to the DNA profiles of potential suspects or individuals in a DNA database.
Paternity and Relationship Testing: DNA fingerprinting is employed to establish biological relationships, such as paternity testing to determine the biological father of a child. By comparing the DNA profiles of the alleged father, child, and mother, it can be determined whether there is a biological relationship.
Missing Persons and Disaster Victim Identification: DNA fingerprinting is used to identify missing persons or victims of mass disasters where traditional identification methods are challenging. DNA samples from the remains or personal belongings of the missing or deceased individuals can be compared to DNA profiles of their relatives to establish their identity.
Immigration and Citizenship Cases: DNA fingerprinting can be utilized in immigration cases to verify family relationships when supporting documents are unavailable or insufficient. By comparing DNA profiles of individuals claiming a biological relationship, such as siblings or parent-child relationships, it can provide evidence for immigration or citizenship applications.
Conservation Biology and Wildlife Forensics: DNA fingerprinting is employed to study and protect endangered species, track illegal wildlife trade, and identify poaching activities. By analyzing DNA samples from animals or their products, such as ivory or skins, the origin of the specimens can be determined.
Medical Research and Genetic Studies: DNA fingerprinting is used in medical and genetic research to study human genetic variation, population genetics, and the inheritance of genetic diseases. By analyzing DNA samples from individuals, researchers can gain insights into the genetic factors associated with certain diseases or traits.
It is important to note that DNA fingerprinting has numerous other applications beyond the ones mentioned above, and its usage continues to expand as technology advances and new applications are discovered.
can dna fingerprinting be used to study evolutionary connections?
Yes, DNA fingerprinting can be used to study evolutionary connections. By comparing DNA fingerprints or genetic profiles of different organisms, scientists can determine the degree of genetic similarity or relatedness between species. This information can be used to trace evolutionary relationships and construct phylogenetic trees, which illustrate the evolutionary history and genetic connections between various organisms.
DNA fingerprinting techniques, such as analyzing specific genetic markers or sequencing DNA regions, allow researchers to compare and analyze the genetic variations and similarities among different species or populations. This information helps in understanding evolutionary patterns, speciation events, genetic diversity, and the relatedness of organisms.
DNA fingerprinting has been instrumental in studying evolutionary connections in various fields, including evolutionary biology, phylogenetics, population genetics, and conservation biology. It provides insights into the genetic relationships between species, the timing of evolutionary events, and the patterns of genetic variation within and between populations.
What sections of dna are used in dna fingerprinting?
In DNA fingerprinting, specific sections of DNA known as genetic markers are used. These genetic markers are regions of the DNA that exhibit high variability between individuals, making them useful for distinguishing one individual from another. The two main types of genetic markers commonly used in DNA fingerprinting are:
Short Tandem Repeats (STRs): STRs are repetitive DNA sequences consisting of short repeating units of 2 to 6 base pairs. These sequences are scattered throughout the genome, including both coding and non-coding regions. The number of repeats at a particular STR locus can vary among individuals, making them highly informative for DNA profiling.
Variable Number Tandem Repeats (VNTRs): VNTRs are also repetitive DNA sequences with longer repeating units compared to STRs, typically consisting of 10 to 100 base pairs. VNTRs are found in specific regions of the genome and can vary greatly in length between individuals. They are less commonly used in modern DNA fingerprinting compared to STRs.
Both STRs and VNTRs are targeted and amplified using polymerase chain reaction (PCR) techniques in the process of DNA fingerprinting. The resulting amplified DNA fragments are then analyzed and compared to generate unique DNA profiles for individuals, which can be used for identification, forensic analysis, and other applications.
What is vntrs dna fingerprinting?
Variable Number Tandem Repeats (VNTRs) were one of the early types of genetic markers used in DNA fingerprinting. VNTRs are regions of DNA where short DNA sequences are repeated in a tandem arrangement, and the number of repeats varies among individuals.
In VNTR analysis, specific VNTR loci are targeted for analysis. The DNA samples are subjected to PCR (Polymerase Chain Reaction), which amplifies the VNTR regions of interest. The PCR products are then separated using gel electrophoresis, which sorts the DNA fragments based on their size. The resulting pattern of DNA fragments is known as a DNA fingerprint or DNA profile.
The analysis of VNTRs involves comparing the number of repeats at different VNTR loci between individuals. Since the number of repeats can vary significantly, VNTR analysis can provide a high level of discrimination between individuals, making it useful for forensic investigations, paternity testing, and population studies.
However, VNTR analysis has certain limitations. The technique requires a larger amount of DNA, and the analysis process can be time-consuming and labor-intensive. Additionally, the interpretation of VNTR profiles can be more challenging compared to newer techniques such as Short Tandem Repeat (STR) analysis, which has largely replaced VNTR analysis in modern DNA fingerprinting due to its advantages of higher sensitivity, specificity, and ease of analysis.
References
- https://geneticeducation.co.in/dna-fingerprinting-definition-steps-methods-and-applications/
- https://www.geeksforgeeks.org/dna-fingerprinting/
- https://www.genome.gov/genetics-glossary/DNA-Fingerprinting#:~:text=DNA%20fingerprinting%20is%20a%20laboratory,that%20are%20unique%20to%20individuals.
- https://www.yourgenome.org/facts/what-is-a-dna-fingerprint/
- https://www.britannica.com/science/DNA-fingerprinting