What is Northern Blotting?
Northern blotting is a laboratory technique used to study gene expression patterns by detecting and analyzing RNA molecules. It is named after its similarity to the Southern blotting technique, which is used for DNA analysis. The Northern blotting method allows researchers to determine the presence, size, and abundance of specific RNA molecules within a sample.
The basic steps involved in Northern blotting are as follows:
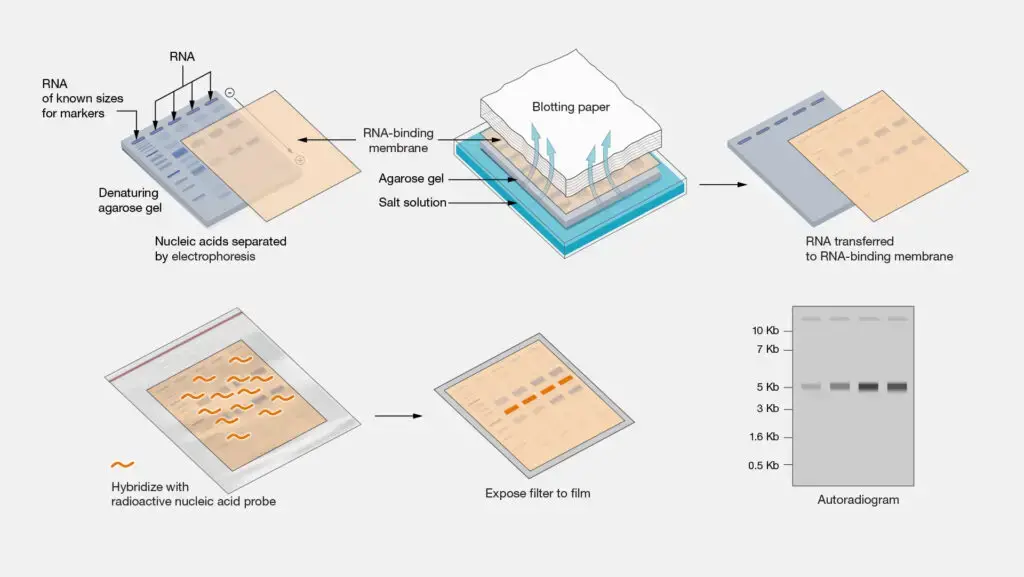
- RNA Extraction: Total RNA is isolated from cells or tissues using methods such as phenol-chloroform extraction or commercial RNA extraction kits. The extracted RNA may include different types of RNA, such as messenger RNA (mRNA), ribosomal RNA (rRNA), and transfer RNA (tRNA).
- RNA Denaturation: The extracted RNA is denatured using heat or chemical denaturants to break the hydrogen bonds and convert the RNA molecules into single-stranded form.
- Gel Electrophoresis: The denatured RNA samples are separated based on their size using gel electrophoresis. Typically, agarose gel is used for Northern blotting, and the RNA samples are loaded into wells in the gel and subjected to an electric field. The RNA molecules migrate through the gel according to their size, with smaller molecules moving faster than larger ones.
- Transfer to Membrane: After gel electrophoresis, the separated RNA molecules are transferred (blotted) from the gel onto a solid support membrane, typically made of nitrocellulose or nylon. This transfer can be achieved through capillary action or by using a vacuum apparatus.
- Hybridization: The transferred RNA on the membrane is then hybridized with a labeled RNA or DNA probe. The probe is a complementary sequence that can specifically bind to the target RNA of interest. The probe is usually labeled with a radioactive isotope (such as 32P) or a non-radioactive marker (such as biotin or digoxigenin) for detection.
- Washing and Detection: After hybridization, the membrane is washed to remove any unbound probe. Detection of the bound probe is achieved by either autoradiography (for radioactive probes) or chemiluminescent/fluorescent methods (for non-radioactive probes). This step allows visualization and quantification of the target RNA bands on the membrane.
What is Southern Blotting?
Southern blotting is a laboratory technique used to detect and analyze specific DNA sequences in a sample. It is named after its inventor, Edwin Southern. The Southern blotting method allows researchers to identify and characterize DNA fragments of interest, such as specific genes, gene mutations, or DNA polymorphisms.
The basic steps involved in Southern blotting are as follows:
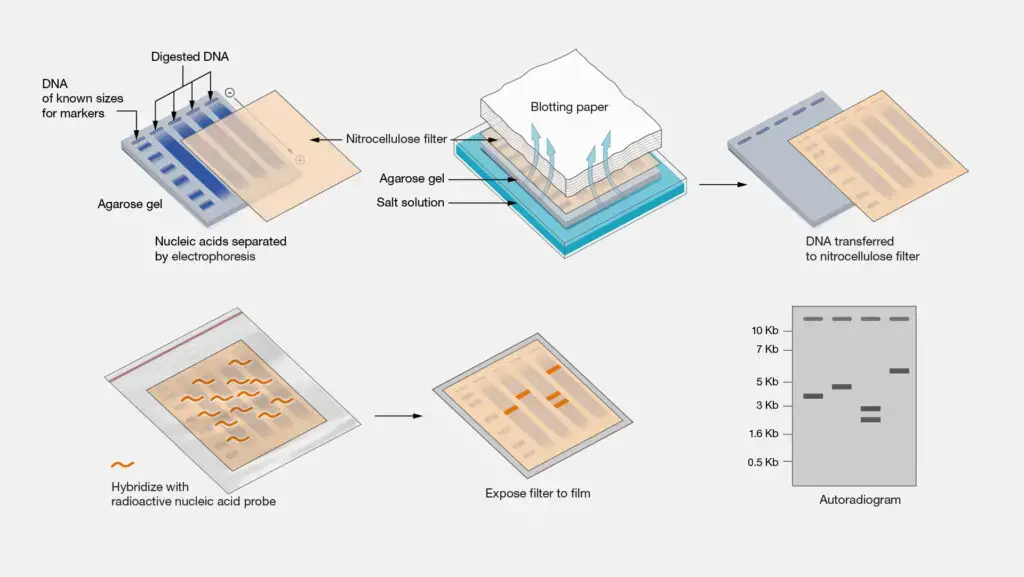
- DNA Extraction: Genomic DNA is isolated from cells or tissues using various extraction methods. The extracted DNA may include both coding and non-coding regions of the genome.
- DNA Digestion: The extracted DNA is digested with restriction enzymes, which are enzymes that recognize specific DNA sequences and cut the DNA at those sites. This step generates DNA fragments of various sizes.
- Gel Electrophoresis: The digested DNA fragments are separated by size using gel electrophoresis. Agarose or polyacrylamide gel is commonly used for Southern blotting. The DNA fragments are loaded into wells in the gel and subjected to an electric field. The fragments migrate through the gel, with smaller fragments moving faster than larger ones.
- Transfer to Membrane: After gel electrophoresis, the separated DNA fragments are transferred (blotted) from the gel onto a solid support membrane, typically made of nitrocellulose or nylon. This transfer can be achieved through capillary action or by using a vacuum apparatus.
- DNA Denaturation: The transferred DNA on the membrane is denatured, usually by treating it with alkali or heat, to convert the double-stranded DNA into single-stranded form.
- Hybridization: The single-stranded DNA on the membrane is then hybridized with a labeled DNA probe. The probe is a complementary sequence that can specifically bind to the target DNA sequence of interest. The probe is typically labeled with a radioactive isotope (such as 32P) or a non-radioactive marker (such as biotin or digoxigenin) for detection.
- Washing and Detection: After hybridization, the membrane is washed to remove any unbound probe. Detection of the bound probe is achieved by either autoradiography (for radioactive probes) or chemiluminescent/fluorescent methods (for non-radioactive probes). This step allows visualization and identification of the target DNA fragments on the membrane.
What is Western Blotting?
Western blotting, also known as immunoblotting, is a widely used laboratory technique for the detection and analysis of specific proteins in a sample. It allows researchers to determine the presence, quantity, and molecular weight of target proteins, as well as investigate protein expression levels, post-translational modifications, and protein-protein interactions.
The basic steps involved in Western blotting are as follows:
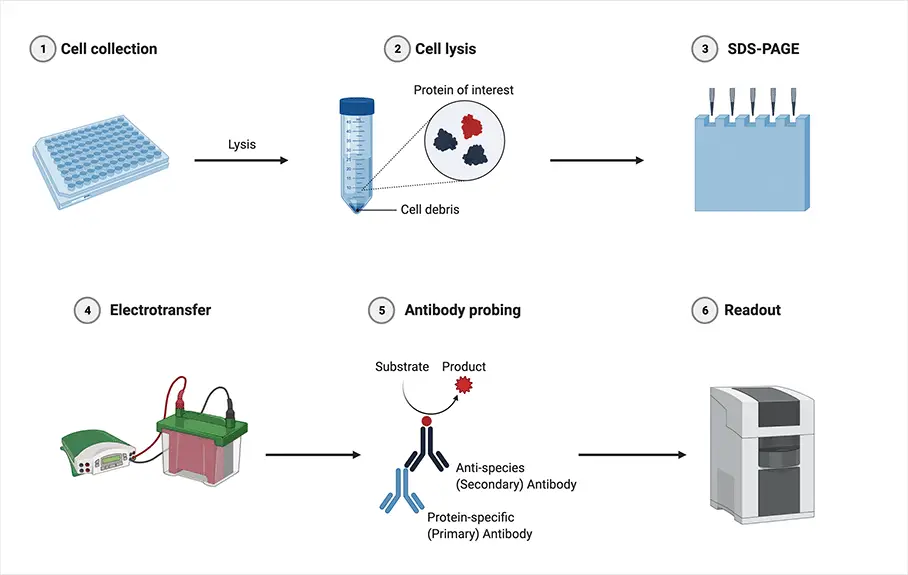
- Protein Extraction and Denaturation: Proteins are extracted from cells or tissues using appropriate lysis buffers to solubilize the proteins. The extracted proteins are then denatured by heating in the presence of a reducing agent, such as β-mercaptoethanol or dithiothreitol (DTT), which breaks the disulfide bonds and unfolds the proteins.
- Protein Separation by SDS-PAGE: The denatured proteins are separated based on their molecular weight using sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The proteins are loaded into wells of an acrylamide gel, and an electric field is applied to migrate the proteins through the gel. The SDS in the gel denatures the proteins further and imparts a negative charge to each protein, causing them to migrate according to their size.
- Protein Transfer to Membrane: After gel electrophoresis, the separated proteins are transferred (blotted) from the gel onto a solid support membrane, commonly made of nitrocellulose or polyvinylidene difluoride (PVDF). This transfer is typically achieved using a technique called electroblotting, in which an electric current is applied to facilitate the transfer of proteins from the gel to the membrane.
- Blocking: The transferred membrane is incubated in a blocking solution, usually a protein-rich solution such as bovine serum albumin (BSA) or non-fat milk, to prevent non-specific binding of antibodies and reduce background noise.
- Antibody Incubation (Primary Antibody): The membrane is then incubated with a primary antibody specific to the protein of interest. The primary antibody recognizes and binds to the target protein. The primary antibody is typically produced by immunizing animals with the purified target protein or through recombinant antibody technologies.
- Washing: After primary antibody incubation, the membrane is washed to remove any unbound antibodies and reduce background noise.
- Antibody Incubation (Secondary Antibody): The membrane is incubated with a secondary antibody conjugated to an enzyme or a fluorescent molecule. The secondary antibody recognizes and binds to the primary antibody, allowing for signal amplification. The secondary antibody is typically raised against the host species of the primary antibody (e.g., anti-mouse secondary antibody for a mouse primary antibody).
- Washing: After secondary antibody incubation, the membrane is washed again to remove any unbound secondary antibodies and reduce background noise.
- Detection: The presence of the target protein is visualized using various detection methods. For enzyme-conjugated secondary antibodies, a substrate specific to the enzyme is added, resulting in the production of a visible signal (e.g., chemiluminescence or color development). For fluorescently labeled secondary antibodies, the signal can be detected using fluorescence imaging systems.
- Analysis: The detected protein bands are analyzed and quantified using appropriate imaging and analysis software. This allows for the determination of protein expression levels, identification of protein isoforms or modifications, and comparison between different samples or experimental conditions.
Difference Between Northern, Southern and Western Blotting
| Feature | Northern Blotting | Southern Blotting | Western Blotting |
|---|---|---|---|
| Target Molecules | RNA | DNA | Proteins |
| Purpose | Study gene expression levels | Identify specific DNA sequences | Detect and analyze specific proteins |
| Sample Preparation | RNA extraction and purification required | DNA extraction and purification required | Protein extraction and denaturation required |
| Electrophoresis Method | Agarose gel | Agarose or polyacrylamide gel | SDS-PAGE (Polyacrylamide gel electrophoresis with SDS) |
| Transfer Method | Capillary or vacuum transfer | Capillary or vacuum transfer | Electroblotting |
| Membrane Type | Nitrocellulose or nylon membrane | Nitrocellulose or nylon membrane | Nitrocellulose or PVDF membrane |
| Probe Types | Radioactive or non-radioactive | Radioactive or non-radioactive | Antibodies |
| Probe Labeling | Typically labeled with radioisotopes (e.g., 32P) | Labeled with radioisotopes (e.g., 32P) or non-radioactive | Enzyme-conjugated or fluorescently labeled antibodies |
| Hybridization | RNA probe hybridizes with complementary RNA sequences | DNA probe hybridizes with complementary DNA sequences | Antibodies bind to specific protein epitopes |
| Detection | Autoradiography or chemiluminescence | Autoradiography or chemiluminescence | Enzymatic or fluorescence detection |
| Sensitivity | Less sensitive than Southern blotting | More sensitive than Northern blotting | Variable sensitivity depending on antibody and detection method |
| Specificity | Detects specific RNA sequences | Detects specific DNA sequences | Detects specific protein targets |
| Application | Gene expression analysis, RNA quantification | DNA fragment analysis, DNA mapping, genetic testing | Protein expression analysis, protein characterization |
| Type of Biological Material | RNA from cells or tissues | DNA from cells or tissues | Proteins from cells or tissues |
| Molecular Weight Information | No molecular weight information | DNA fragment size information | Protein size information |
FAQ
What is the primary difference between Northern, Southern, and Western blotting?
The key difference lies in the type of biomolecule being detected. Northern blotting analyzes RNA molecules, Southern blotting detects DNA sequences, and Western blotting examines proteins.
What are the differences in sample preparation for these techniques?
Northern and Southern blotting require extraction and purification of RNA and DNA, respectively. In contrast, Western blotting involves protein extraction and denaturation.
How do these techniques differ in terms of gel electrophoresis?
Northern and Southern blotting commonly use agarose gel electrophoresis, while Western blotting employs SDS-PAGE (Polyacrylamide gel electrophoresis with SDS).
What are the membrane types used in these blotting methods?
Nitrocellulose or nylon membranes are typically used in both Northern and Southern blotting. Western blotting commonly employs nitrocellulose or PVDF (Polyvinylidene difluoride) membranes.
What types of probes are used in each technique?
Northern and Southern blotting use RNA or DNA probes, respectively, which are typically labeled with radioactive or non-radioactive markers. Western blotting employs antibodies as probes.
How do the hybridization steps differ in these blotting techniques?
In Northern and Southern blotting, the RNA or DNA probe hybridizes with complementary RNA or DNA sequences, respectively. In Western blotting, antibodies specifically bind to target proteins.
What are the main detection methods employed in these techniques?
Northern and Southern blotting commonly use autoradiography or chemiluminescence for detection. Western blotting uses enzymatic or fluorescence-based detection methods.
Are there differences in sensitivity among these techniques?
Generally, Southern blotting is more sensitive than Northern blotting due to the stability of DNA. Western blotting sensitivity can vary depending on the specific antibodies and detection methods used.
What are the primary applications of each blotting technique?
Northern blotting is commonly used for gene expression analysis and RNA quantification. Southern blotting is applied in DNA fragment analysis, DNA mapping, and genetic testing. Western blotting is used for protein expression analysis and characterization.
Can these techniques provide information about molecular weight?
While Southern blotting can provide DNA fragment size information, Northern blotting does not offer molecular weight details. Western blotting can give insights into protein size based on migration in SDS-PAGE gels.
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.