What is gene/genome editing?
Gene or genome editing refers to the deliberate and precise modification of an organism’s genetic material. This advanced biotechnological process allows for the targeted alteration of DNA sequences within a genome, facilitating the correction of genetic anomalies or the introduction of specific traits.
Historically, the concept of gene editing traces its roots back to the late 1970s, marked by the groundbreaking work of Herb Boyer and Stanley Cohen, who successfully engineered antibiotic-resistant bacteria. However, it wasn’t until 2012 that the field witnessed a revolutionary breakthrough with the discovery of the CRISPR-Cas9 system. Originating from a bacterial immune defense mechanism, CRISPR-Cas9 offers a highly efficient and targeted approach to gene editing. This discovery, led by researchers from the University of California, transformed the landscape of genetic engineering.
Interestingly, the journey towards this innovation began much earlier. In 1982, scientists engineered bacteria to produce human insulin, a synthetic version that later received FDA approval and commercial distribution. Following this success, the path was paved for the development of genetically modified organisms, including the Flvr Savr tomato and certain cotton species.
The gene editing process is methodical and involves several crucial steps:
- Target Identification: Pinpointing the specific genomic site requiring modification.
- Healthy Copy Design: Creating a corrected version of the targeted DNA sequence.
- Guide RNA Design: Crafting RNA molecules that guide the editing machinery to the desired location.
- Enzymatic Action: Utilizing nucleases, often termed “molecular scissors,” to cleave the erroneous DNA segment.
- DNA Insertion: Introducing the corrected DNA sequence at the target site.
- Repair: Leveraging the cell’s inherent DNA repair mechanisms to mend the edited site.
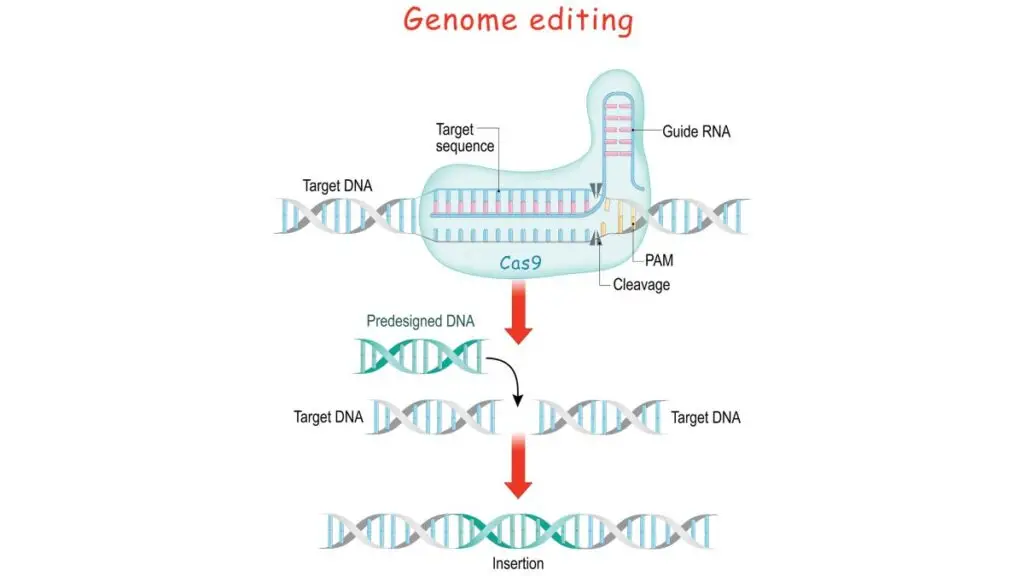
The central player in this process is the guide RNA or gRNA, which binds to the target DNA sequence. Assisted by an enzyme, typically a nuclease, the DNA is then cleaved at the specified location. Following this, the cell’s intrinsic repair machinery rectifies the break, either by natural processes or with the insertion of a desired DNA fragment.
While gene editing has been successfully applied to various organisms like bacteria, yeast, and mice, its application in humans remains controversial due to ethical considerations, especially concerning human embryos.
In essence, gene editing represents a monumental leap in biotechnology, offering unprecedented control over genetic material. As depicted in the accompanying illustration, this intricate process holds immense potential for advancing medical science and addressing genetic disorders.
What is the need for gene-editing?
- Gene editing emerges as a pivotal tool in the realm of biotechnology, addressing the pressing challenges posed by genetic disorders. These disorders, often lethal and predominantly incurable, have long perplexed the scientific community. With the groundbreaking discovery of DNA in 1953, the intricate mechanisms underlying the inheritance of diseases were elucidated. Chromosomes, the cellular structures housing DNA, play a central role in transmitting genetic information across generations.
- This hereditary transmission is not limited to beneficial traits alone. Mutations, or alterations in the DNA sequence, can also be passed down, leading to genetic abnormalities in subsequent generations. Such mutations can manifest as genetic disorders, impacting the health and well-being of affected individuals.
- Gene editing offers a promising solution to this challenge. By enabling precise modifications at the genetic level, gene editing can target and rectify these mutations, preventing their propagation to future generations. In essence, this technology holds the potential to eliminate the mutated segments of the genome, thereby mitigating the risk of inherited genetic disorders. In a world where genetic anomalies pose significant health threats, the need for gene editing becomes paramount, offering hope for a future free from the burden of genetic diseases.
Importance of gene editing
Gene editing stands as a cornerstone in modern biotechnology, underpinning numerous scientific and medical advancements. Its significance is multifaceted, encompassing a range of applications across various organisms.
- Broad Applicability: Gene editing is versatile, allowing modifications in the genomes of diverse organisms. While its utility is evident in model organisms such as bacteria, yeast, and mice, its potential extends far beyond these entities, offering insights into the genetic underpinnings of a plethora of life forms.
- Elucidating Gene Expression: A pivotal aspect of gene editing lies in its capacity to study gene expression. By manipulating specific genes, researchers can discern their roles, functions, and interactions within an organism, thereby deepening our understanding of genetic regulation and its implications.
- Rectifying Genetic Anomalies: One of the most transformative applications of gene editing is its ability to target and eliminate mutated genes within an organism’s genome. Such precision not only holds promise for addressing genetic disorders but also paves the way for the development of novel genetic variations.
- Tailoring Organismal Traits: Gene editing empowers scientists to modify the characteristics of organisms deliberately. Whether it’s enhancing a crop’s resistance to pests or altering an animal’s physical attributes, this technology offers unparalleled control over an organism’s genetic makeup.
- Economic Relevance: Beyond its scientific implications, gene editing has substantial economic ramifications. By facilitating the creation of genetically modified organisms tailored for specific purposes, it can drive innovations in agriculture, medicine, and other sectors, leading to economic growth and sustainability.
In essence, the importance of gene editing transcends mere scientific curiosity. It holds the key to unlocking myriad possibilities, from advancing medical research to fostering sustainable development, underscoring its indispensable role in the future of science and society.
Types of gene-editing
Gene editing, a transformative biotechnological tool, employs specialized enzymes known as nucleases to make precise modifications in the DNA of an organism. These nucleases, often referred to as “molecular scissors,” are pivotal in facilitating targeted genetic alterations. Based on the type of nuclease utilized, gene editing can be categorized into several distinct techniques. Here, we delve into the three primary methods that have gained prominence in the realm of genetic engineering:
- ZFNs (Zinc Finger Nucleases): ZFNs are engineered proteins that combine the DNA-binding capabilities of zinc finger proteins with the DNA-cleaving activity of the FokI nuclease. By designing specific zinc finger domains, ZFNs can be tailored to target unique DNA sequences, enabling precise genetic modifications.
- TALEN (Transcription Activator-Like Effector Nucleases): TALENs are another class of engineered nucleases that merge the DNA-binding domain of transcription activator-like effectors (TALEs) with the FokI nuclease domain. Similar to ZFNs, TALENs can be customized to recognize and bind to specific DNA sequences, facilitating targeted gene editing.
- CRISPR-Cas9 (Clustered Regularly Interspaced Short Palindromic Repeats): Arguably the most revolutionary gene-editing tool in recent times, CRISPR-Cas9 is derived from a bacterial defense mechanism against viruses. The system employs RNA molecules, designed to match target DNA sequences, in conjunction with the Cas9 nuclease enzyme. Upon binding to the target DNA, Cas9 induces a cut, allowing for the addition, deletion, or replacement of genetic material.
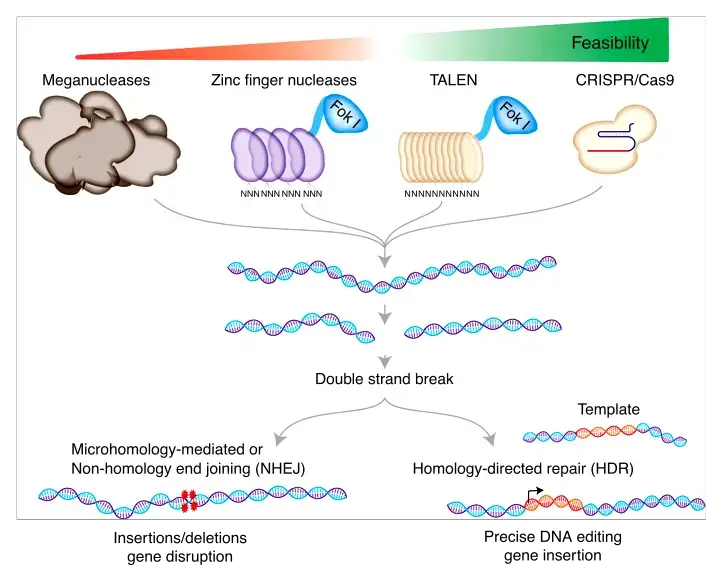
What is TALENs?
- Transcription activator-like effector nucleases (TALENs) are DNA-binding proteins with an array of 33 or 34 repeating amino acids.
- TALENs are synthetic restriction enzymes created by combining the DNA-cutting domain of a nuclease with TALE domains, which may be programmed to identify a specific DNA sequence.
- These fusion proteins serve as readily targetable “DNA scissors” for gene editing applications, enabling targeted genome modifications in living cells, including sequence insertion, deletion, repair, and replacement.
- The DNA binding domains, which may be engineered to bind any desired DNA sequence, are derived from TAL effectors, which are DNA-binding proteins excreted by the plant-pathogenic Xanthomanos spp.
- TAL effectors are composed of repetitive domains, each containing a highly conserved sequence of 34 amino acids, and they detect a single DNA nucleotide within the target region.
- The nuclease is capable of creating double strand breaks at the target location, which can be repaired by error-prone non-homologous end-joining (NHEJ), resulting in gene disruptions caused by the introduction of tiny insertions or deletions. Except for the so-called repeat variable di-residues (RVDs) at amino acid positions 12 and 13, each repeat is conserved.
- RVDs are responsible for determining the DNA sequence to which the TALE will attach. This clear one-to-one correlation between TALE repeats and the appropriate DNA sequence simplifies the process of constructing repeat arrays to identify novel DNA sequences.
- These TALEs can be joined with the catalytic domain of the DNA nuclease FokI to produce a transcription activator-like effector nuclease (TALEN). Combining specificity with activity, the resultant TALEN constructions provide tailored sequence-specific nucleases that bind and cleave DNA sequences only at predetermined places.
- The TALEN target recognition system utilises a code that is simple to predict. In part because to the length of their 30+ base pair binding site, TAL nucleases are unique to their target. Within a range of six base pairs, TALEN may modify any single nucleotide in the whole genome.
- Similar to engineered zinc finger nucleases, TALEN constructions have three advantages in targeted mutagenesis: (1) improved DNA binding specificity, (2) lesser off-target effects, and (3) faster assembly of DNA-binding domains.
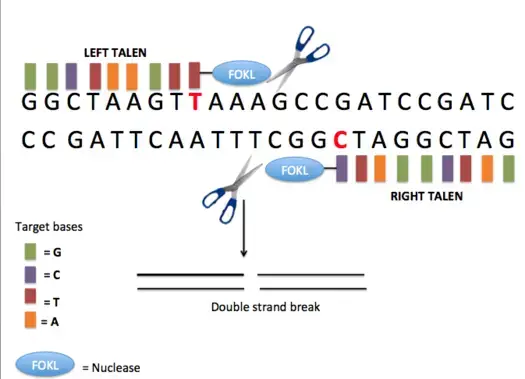
What is ZFNs?
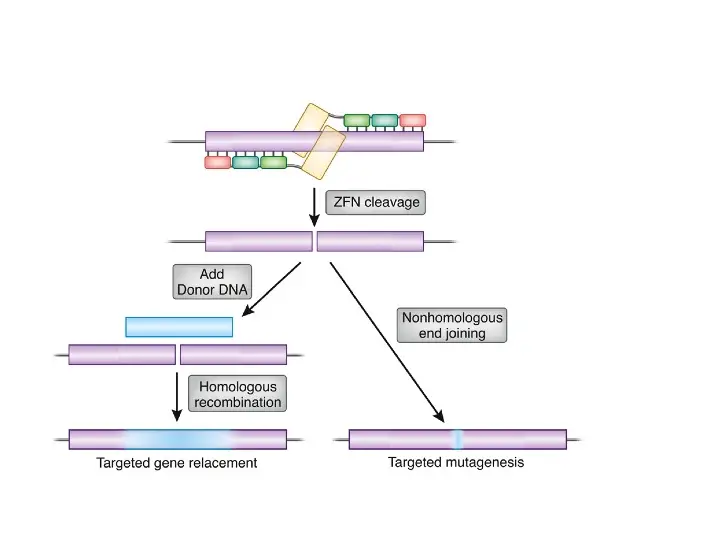
- Zinc-finger nucleases (ZFNs) are synthetic restriction enzymes made by combining a zinc finger DNA-binding domain with a DNA-cleavage domain.
- The ability to build zinc finger domains to target specific DNA sequences enables zinc-finger nucleases to target unique sequences within complicated genomes.
- These reagents can be used to precisely modify the genomes of higher species by utilising endogenous DNA repair machinery.
- In addition to CRISPR/Cas9 and TALEN, ZFN is a significant technology for genome editing.
- Zinc finger nucleases are beneficial for manipulating the genomes of numerous plants and animals, such as arabidopsis, tobacco, soybean, corn, Drosophila melanogaster, and Candida albicans. elegans, Platynereis dumerilii, sea urchin, silkworm, zebrafish, frogs, mice, rats, rabbits, pigs, and cattle, as well as many mammalian cell types.
- Zinc finger nucleases have also been employed in a mouse model of haemophilia, and a clinical experiment indicated that CD4+ human T-cells with the CCR5 gene damaged by zinc finger nucleases are safe as a potential HIV/AIDS treatment.
- ZFNs are also employed to produce isogenic human illness models, a new generation of genetic disease models.
What is CRISPR-Cas9?
- CRISPR-Cas9 is a genome-editing technique that is generating excitement within the scientific community. It is faster, less expensive, and more precise than earlier DNA editing approaches, and it has a wide range of possible uses.
- CRISPR-Cas9 is an innovative technology that allows geneticists and medical researchers to edit portions of the genome by removing, inserting, or modifying DNA sequence segments.
- In common usage, “CRISPR” (pronounced “crisper”) is an abbreviation for “CRISPR-Cas9.” CRISPRs are specific sequences of DNA, and the protein Cas9 — where Cas stands for “CRISPR-associated” — is an enzyme capable of severing DNA strands.
- Adapted from the natural defence mechanisms of bacteria and archaea, a category of relatively primitive single-celled microorganisms, CRISPR technology is a form of gene editing. Viruses are repelled by these species using CRISPR-derived RNA, a molecular cousin to DNA, and different Cas proteins.
- In order to thwart attacks, organisms sever the DNA of viruses and then store fragments of that DNA in their own genome to be deployed as a weapon if the viruses attack again.
- When the components of CRISPR are transferred into other, more complex organisms, these components can then manipulate genes, a process known as “gene editing.” No one knew what this process looked like until 2017, when a team of researchers led by Mikihiro Shibata of Kanazawa University in Japan and Hiroshi Nishimasu of the University of Tokyo revealed for the first time what a CRISPR looks like in action.
- It is currently the easiest, most adaptable, and most accurate approach of genetic manipulation, and is consequently generating a buzz in the scientific community.
- Comparable to an immune system, certain bacteria have an innate gene-editing mechanism similar to CRISPR-Cas9 that they use to respond to invading pathogens such as viruses.
- Using CRISPR, the bacteria remove portions of the virus’ DNA while retaining a portion of it to enable them recognise and defend against the virus in the future.
- Scientists modified this technique so that it could be employed in various animal cells, including those of humans and mice.
- What have scientists discovered about genetics throughout the years? and gene function by investigating the consequences of DNA mutations.
- If it is possible to alter a gene in a cell line or an entire organism, it is possible to examine the effect of this alteration in order to determine the function of the gene.
- Long ago, geneticists caused mutations using chemicals or radiation. However, there was no method to control where mutations occur in the genome.
- Scientists have been employing “gene targeting” for a number of years to induce alterations in specific locations of the genome by removing or inserting complete genes or single nucleotides.
- Traditional gene targeting has been very useful for researching genes and genetics, but creating a mutation is time-consuming and costly.
- Several ‘gene editing’ technologies, including CRISPR-Cas systems, transcription activator-like effector nucleases (TALENs), and zinc-finger nucleases, have recently been created to improve gene targeting techniques (ZFNs).
- The CRISPR-Cas9 system is currently the most efficient, cost-effective, and trustworthy method for ‘editing’ genes.
Components Of Crispr
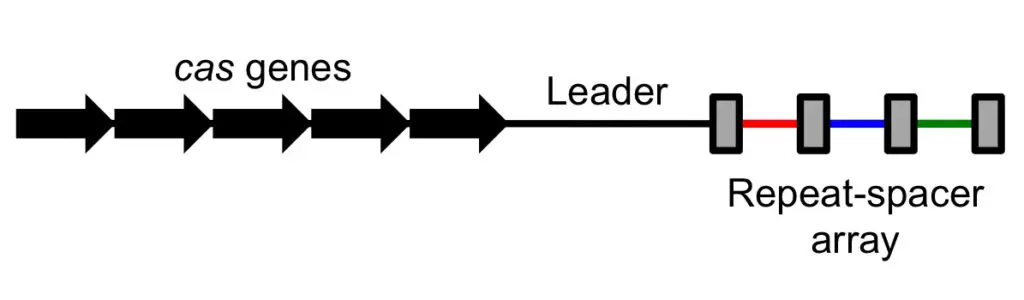
1. CRISPRs
- CRISPR is an acronym for “clusters of regularly interspaced short palindromic repeats” and it refers to a section of DNA composed of short, repeating sequences with so-called “spacers” placed between each repeat.
- Repeats in the genetic code refer to the ordering of rungs within the spiral ladder of a DNA molecule. Each rung comprises two chemical bases linked together: adenine (A) is linked to thymine (T), and guanine (G) is linked to cytosine (C) (C).
- According to the Max Planck Institute, in a CRISPR area, these bases are repeated in the same order multiple times, forming what are known as “palindromic” sequences.
- In a palindromic sequence, the bases on one side of the DNA ladder match those on the opposite side when they are read in opposite directions.
- A highly simple palindromic sequence might seem like follows:
- Side 1 – GATC
- Side 2 – CTAG
- Short palindromic repeats are found throughout CRISPR sections of DNA, with each repeat flanked by “spacers.” Bacteria steal these spacers from viruses that have attacked them, so incorporating a portion of viral DNA into their own genome.
- These spacers function as a memory bank, allowing the bacteria to recognise the viruses should they ever strike again. You may also compare spacers to “Wanted” posters, as they provide a snapshot of the criminals, allowing them to be readily identified and brought to punishment.
- This procedure was originally experimentally proved by Rodolphe Barrangou and a team of researchers at Danisco, a manufacturer of food additives.
- The researchers utilised Streptococcus thermophilus bacteria, which are often found in yoghurt and other dairy cultures, as their model in a 2007 paper published in the journal Science, according to the Joint Genome Institute, part of the U.S. Ministry of Energy.
- After a viral infection, the bacteria were observed to incorporate additional spacers into their CRISPR regions. In addition, the DNA sequence of these spacers was identical to that of portions of the virus genome.
- Additionally, the team removed the spacers and inserted fresh viral DNA sequences in their place. In this manner, the researchers were able to modify the bacteria’s resistance to a particular virus, proving the significance of CRISPRs in regulating bacterial immunity.
2. CRISPR RNA (crRNA)
- CRISPR sections of DNA serve as a sort of memory bank for viral information; however, for this information to be useful elsewhere in the cell, it must be replicated, or “transcribed,” into RNA.
- Unlike DNA sequences, which remain lodged within the DNA molecule, this CRISPR RNA (crRNA) can roam throughout the cell and collaborate with proteins, namely the molecular scissors that snip viruses to pieces.
- RNA varies from DNA in that it consists of a single strand, as opposed to two, giving it the appearance of half a ladder.
- To generate an RNA molecule, one portion of CRISPR serves as a template, and proteins known as polymerases construct an RNA molecule that is “complementary” to the template, meaning that the bases of the two strands fit together like puzzle pieces. A G in the DNA molecule, for instance, would be translated as a C in the RNA.
- According to a 2014 assessment published in the journal Science by Jennifer Doudna and Emmanuelle Charpentier, each snippet of CRISPR RNA comprises a copy of a repeat and a spacer from a CRISPR region of DNA.
- The crRNA interacts with the Cas9 protein and another type of RNA, known as “trans-activating crRNA” or tracrRNA, to aid bacteria in their fight against viruses.
3. Cas9
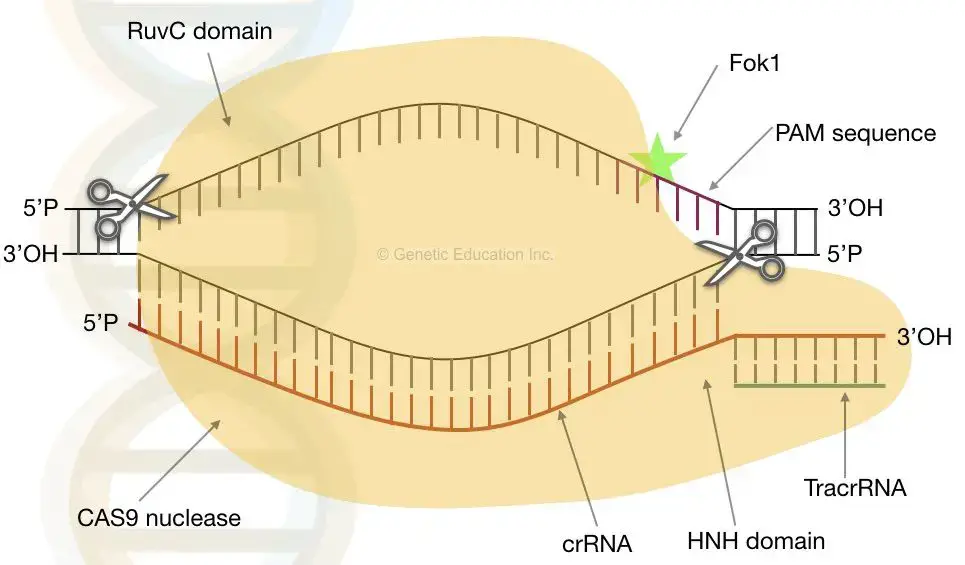
- Cas9 is an enzyme capable of severing foreign DNA. The protein attaches to crRNA and tracrRNA, which together direct Cas9 to the spot on the viral DNA strand where it will make its cut.
- Cas9 will cut through target DNA that is complementary to a 20-nucleotide length of the crRNA, where a “nucleotide” is a single-base DNA building block.
- Using two distinct sections or “domains” on its structure, Cas9 cuts both strands of the DNA double helix, creating a “double-stranded break,” according to a 2014 research published in Science.
- Cas9 is equipped with a safety mechanism that prevents it from randomly slicing a genome. PAMs, or protospacer adjacent motifs, are short DNA sequences that function as tags that are located adjacent to the target DNA sequence.
- If there is no PAM adjacent to the target DNA sequence, Cas9 will not cut. This is one possible explanation for why Cas9 never attacks the CRISPR region in bacteria, according to a 2014 Nature Biotechnology review (opens in new tab).
CRISPR-CAS9 gene editing system
- Some bacteria and archaea contain CRISPR- Clustered Regularly Interspaced Short Palindromic Repeats. It is a component of the natural defensive mechanism of bacteria. The RNA generated from CRISPR protects bacteria against invading viruses and phages.
- The nuclease, CAS9 protein cuts and destroys the DNA strand of invading infections.
- In 1980, the gene sequences for CRISPR were identified in E. coli, although their function was unknown at the time. Later, in 2007, Barrangou et al. reported their function in the bacteria’s adaptive immune system.
CRISPR-CAS9 in Bacteria
- Throughout the viral infection, the virus injects its DNA through the bacterial cell wall.
- The DNA of the phage is integrated into the host genome and then transcribed and translated.
- The viral protein is generated from the bacterial DNA.
- To escape phage infection, bacteria have created a distinctive defence mechanism known as a CRISPR-CAS9-based defence mechanism.
- During the viral invasion, the bacteria added reference DNA sequences into the palindromic repeats that create CRISPR. Combining bacterial palindromic repeat sequences and viral spacer DNA creates CRISPR.
- The spacer DNA is transcribed into the RNA called crRNA, which directs viral genome binding at a precise site. The DNA-RNA heteroduplex is formed at the invasion site.
- The bacteria also produced a number of short tracrRNA (trans-activating crRNA), which are partially complementary to the crRNA.
- The tracrRNA facilitates the maturation of numerous pre-crRNA into mature crRNA.
- gRNA or guided RNA is the junction of crRNA and tracrRNA that directs the nuclease to cut the target sequence.
- This gRNA is recognised by CAS9, which binds to the site of the heteroduplex. Two unrelated domains of a nuclease (CAS9 nuclease) designated a RuvC-like domain and HNH domains lyses the phage dsDNA after it is suitably positioned.
- Throughout this procedure, the CRISPR sequences serve as a target for nuclease to cut the DNA. The DNA of the virus or phase cannot reproduce and destroy.
- The RuvC domain severing the non-homologous domain, whereas the HNH domain severing the homologous domain (homologous to gRNA).
- CRISPR and CAS9 protein work in concert to eliminate the invading pathogen or virus. This theory raises the question, “Does it cut its own DNA? The response is “Yes”.
- The viral DNA sequence is also found in CRISPR. Therefore, it is possible that CRISPR-CAS9 detects these spacer sequences as foreign DNA and edits them accordingly. Actually, it does not occur because of a pattern known as a PAM!
- The photo spacer-associated motif (PAM) aids in gRNA recognition by CAS9. The nuclease is situated close to the PAM domain. It binds to the 3′-OH terminus of gRNA.
- This will prompt CAS9 to begin cleaving the DNA. CAS9 detects the PAM-gRNA complex on double-stranded DNA and catalyses the process. Lack of PAM in the DNA of bacteria protects them. It is a type of safety mechanism designed by bacteria themselves.
- PAM safeguards bacterial DNA against its own nuclease activity.
- CRISPR are the DNA sequences that are composed of spacers and repeats. The spacers are present throughout the CRISPR between the short palindromic repeats. Different spacer sequences are extracted from various viruses to aid bacteria during future pathogen attacks.
- These sequences contain information that bacteria employ to eliminate pathogens. But how can we put it to use?
- We are able to construct synthesised gRNA that directs CAS9 to the desired target site, allowing us to conduct a variety of genome-editing experiments.
CRISPR-CAS9 in genetic engineering
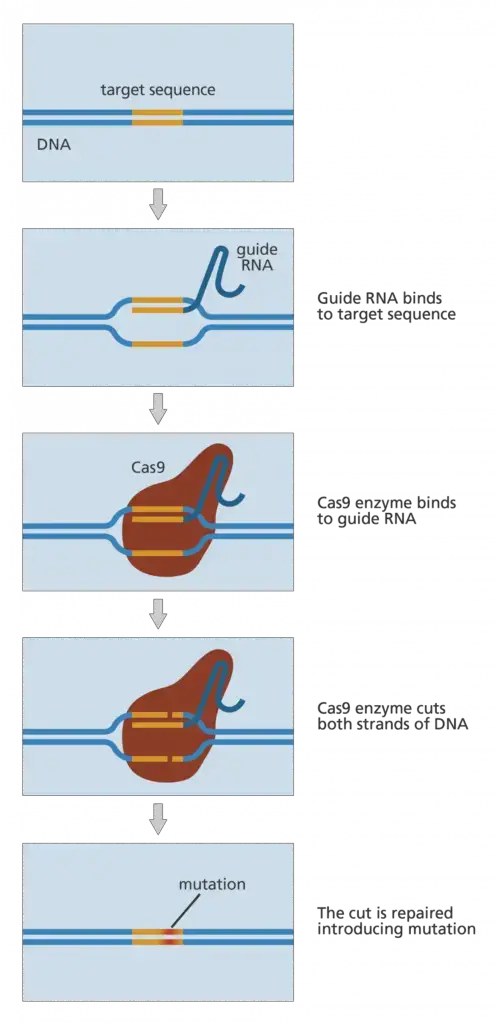
- Guided RNA and the dimeric CAS9 protein are the only prerequisites for gene editing.
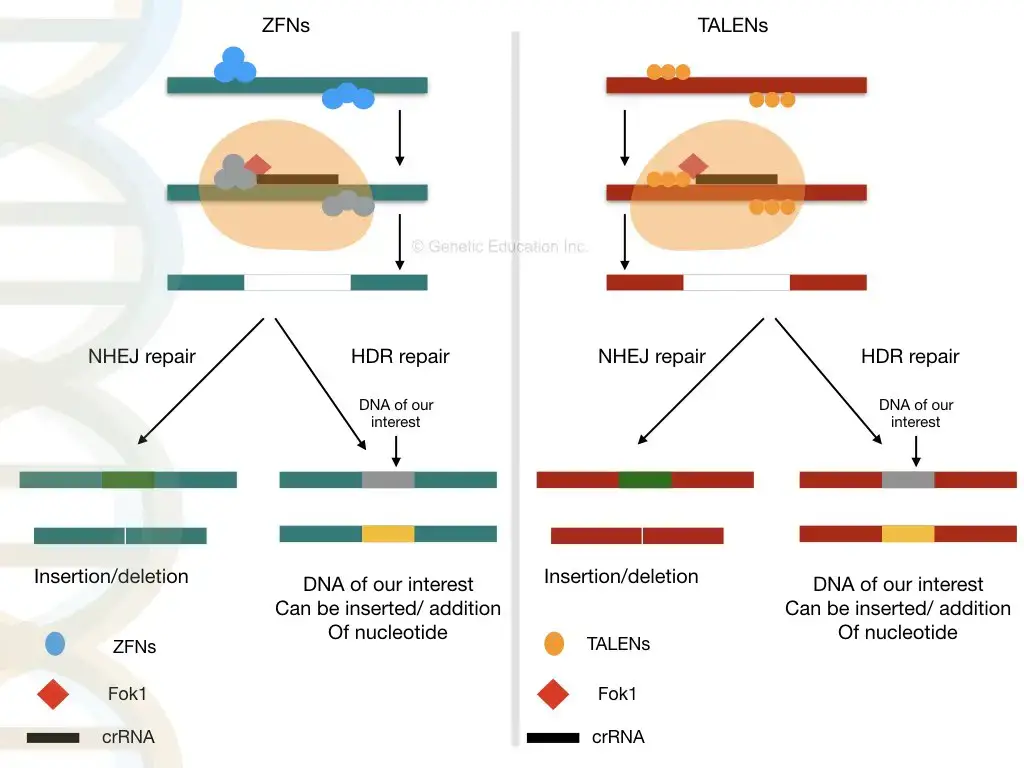
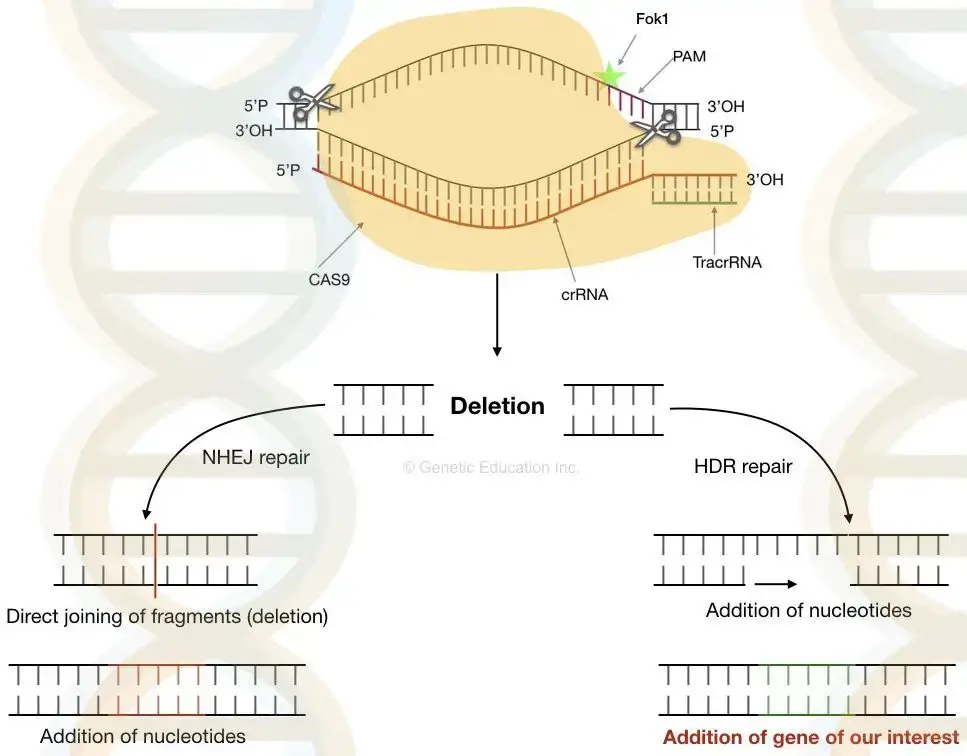
- Once the nuclease has cleaved the DNA at the designated place, it generates the double-stranded cut. Now, the cell’s natural DNA repair system utilises either non-homologous end-joining or Homologous direct repair to close the gap.
- The entire system functions as a result of double-stranded break repair.
Non-homologous end-joining
- In this form of DNA repair process, the gap is directly closed by uniting two non-homologous strands that are opposite one another. Several essential pieces of genetic information may be lost in NHEJ because the whole DNA fragment is missing.
- This DNA fragment will never be present in the subsequent cell division, causing a mutation in the genome. The condition becomes fatal for the organism if some of its essential genes are eliminated.
Homologous direct repair
- In homologous repair, the gaps are not directly closed. Homologous direct repair follows the recombination mechanism. Here, the nucleotides are inserted at the cut spot depending on the data from the previous replication.
- The cell employs single-stranded DNA as a template for the insertion of additional nucleotides throughout this process. Now is the moment to introduce our desired single-stranded DNA, which is generated spontaneously by the cell throughout successive cell divisions.
- By using non-homologous end joining, we can eliminate defective DNA while homologous direct repair can insert DNA.
- CRISPR-CAS9 is utilised in molecular genetics and genetic engineering as a gene-editing technology. On the basis of the activity of CAS9, three distinct gene-editing systems are hypothesised. The following are the three types of systems:
- CAS9 with cleavage activity only
- Full CAS9 nuclease activity
- CAS without the activity of cleavage.
- In the first approach, the CAS9 encoding gene is used to generate the cas9D10A mutant. This mutant is incapable of severing both strands of DNA. Instead, cas9D10A recognises the gRNA and only cleaves one strand of the target DNA.
- What are the advantages of this mutant? In fact, it improves the accuracy of gene editing. It permits only high-fidelity homologous direct repair of the single-stranded gap, hence decreasing the likelihood of deletion-insertion mutations in the gnome.
- In brief, the cas9D10A variation prohibits non-homologous end-joining.
- The second mechanism is CRISPR-CAS9 functioning normally. None of the sequences for CAS9 proteins are altered. We have already described the CRISPR-CAS9 technology and how standard CAS9 operates.
- The third system relies on the dCas9 gene. dCas9 is often referred to as nuclease-deficient CAS9 or dead Cas. Some mutations induced artificially into the RuvC and HNH domains inhibit the function of nuclease.
- The nuclease-deficient CAS9 cannot cut the target sequence; it can only bind to it. With the aid of several effector domains, this permits the silencing or activation of a certain gene.
How does the CRISPR CAS9 System Distinguish Self vs Nonself DNA?
The CRISPR-Cas9 system, a revolutionary genome editing tool, has an intrinsic ability to differentiate between self and non-self DNA, ensuring its precision in targeting foreign genetic elements while preserving the host’s genetic integrity. This distinction is crucial for bacteria to defend against invading pathogens without inadvertently damaging their own DNA. Here, we delve into the mechanisms that underpin this remarkable selectivity.
Mechanisms of Self vs. Non-Self Discrimination:
- Replication Speed Discrepancy: Research by Qimron S and colleagues suggests that the proteins Cas1 and Cas2, integral to spacer acquisition, predominantly target rapidly replicating DNA, typically characteristic of invading DNA. This suggests that foreign DNA replicates more swiftly than bacterial DNA. However, replication speed alone doesn’t account for the entire discrimination process.
- Chi-Sequences as Self-Markers: Certain regions in bacterial DNA, termed “chi-sequences,” act as markers of self. These sequences deter the CRISPR system from acting upon them, ensuring that the Cas proteins predominantly target “chi-less” sequences, which are typically viral in origin.
- PAM Sequences in Invading DNA: The presence of Protospacer Adjacent Motifs (PAM) near the RNA-DNA junction serves as a marker for non-self DNA. These short sequences, ranging from 2 to 6 nucleotides, are recognized by the crRNA-Cas complex, aiding in the distinction between host and foreign DNA. Mutations within the PAM region can, however, disrupt this recognition process.
- 5’ Tag in Type III CRISPR System: In the Type III CRISPR system, a unique 5’ tag present on mature crRNA aids in non-self recognition. This tag remains uninvolved in base pairing with target sequences, facilitating the identification and subsequent degradation of foreign nucleic acids.
- Phage or Plasmid Copy Numbers: Wissman J’s research indicates that as the process of spacer acquisition intensifies, the number of phage DNA or plasmid copies within the host cell escalates, signaling the presence of non-self DNA.
- Cooperation with Restriction Modification System: The Restriction Modification system, another bacterial defense mechanism, complements the CRISPR-mediated immune response. Restriction enzymes cleave foreign DNA at specific recognition sites, providing raw material for the CRISPR system to update its genetic memory. Methylation patterns on host DNA further assist the CRISPR system in distinguishing between self and non-self DNA.
- PFS in Type VI System: In systems targeting RNA, the Protospacer Flanking Sequence (PFS) plays a pivotal role in interference and signals the destruction of phage DNA.
The CRISPR-Cas9 system’s ability to discern between self and non-self DNA is underpinned by a combination of intrinsic and extrinsic markers, ensuring its precision in targeting foreign invaders. This intricate balance safeguards the bacterial genome while providing a robust defense against external threats. As our understanding of these mechanisms deepens, it underscores the sophistication and adaptability of the CRISPR-Cas9 system in the realm of genome editing.
CRISPR-CAS9 in the Treatment of Ataxia Type 2
Spinocerebellar Ataxia Type 2 (SCA2) is a genetically inherited neurodegenerative disorder characterized by an abnormal expansion of the CAG triplet repeat within the ATXN2 gene. This expansion leads to the production of a faulty ataxin-2 protein, which is believed to be the primary cause of the disease’s symptoms, including muscle coordination issues, jerky movements, and speech difficulties. The progressive nature of SCA2 and its autosomal dominant inheritance pattern make it a prime candidate for genetic interventions. One such promising approach is the use of the CRISPR-Cas9 system.
Understanding SCA2 at the Genetic Level
SCA2 arises from the expansion of CAG repeats in the ATXN2 gene, located on chromosome 12. While a normal ATXN2 gene contains up to 22 CAG repeats, individuals with more than 32 repeats are at risk of developing the disease. The overproduction of the ataxin-2 protein due to this expansion is believed to cause the neurological manifestations of SCA2. The disease follows a pattern of genetic anticipation, where the severity increases with each generation.
The Promise of CRISPR-Cas9 in Treating SCA2
The CRISPR-Cas9 system offers a precise method for editing genes at specific locations. This system comprises three main components: the Cas9 protein, CRISPR sequences, and a guide RNA (sgRNA). The sgRNA is designed to be complementary to the target DNA sequence, guiding the Cas9 protein to the desired location for gene editing.
Research by Bryan S, Beverly D, and Alejandro M (2020) highlighted the potential of CRISPR-Cas9 in treating SCA2. Their work demonstrated that targeting exon 1 of the ATXN2 gene effectively silenced its expression, reducing the levels of the faulty ataxin-2 protein in cell lines. This approach, if translated to clinical settings, could potentially halt or even reverse the progression of SCA2.
Furthermore, studies on other types of ataxias, such as the work by Salvatori F & Pappada M et al. (2020) on Spinocerebellar Ataxia Type 1, have shown the versatility of the CRISPR-Cas9 system in targeting different genes associated with ataxias.
Challenges and Considerations
While the potential of CRISPR-Cas9 in treating SCA2 is evident, several challenges remain. Most of the studies conducted so far have been on isolated cell lines or animal models, and the translation to human patients is still in its infancy. Moreover, the efficiency of CRISPR-Cas9 in gene silencing is still under debate, especially when compared to other methods like RNA interference.
Additionally, the precision of CRISPR-Cas9, while remarkable, is not absolute. Off-target effects, where unintended regions of the genome are edited, can lead to unforeseen consequences. The cost, complexity, and potential side effects of the procedure also raise concerns about its widespread application.
The CRISPR-Cas9 system holds significant promise in the realm of genetic disorders, including Spinocerebellar Ataxia Type 2. While preliminary studies have shown its potential in silencing the faulty ATXN2 gene, more comprehensive research and clinical trials are needed to establish its safety and efficacy in patients. As the field of gene editing continues to evolve, it is hoped that CRISPR-Cas9 will play a pivotal role in the treatment of SCA2 and other neurodegenerative disorders.
Importance of PAM Sequence (Protospacer Adjacent Motif) in CRISPR System
The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) system, a revolutionary tool in the realm of genetic engineering, relies heavily on the Protospacer Adjacent Motif, commonly known as PAM. This motif is a short sequence, ranging from 2 to 6 nucleotides (nt) in length, situated adjacent to the protospacer region on the invasive DNA.
Characteristics and Role of PAM Sequence:
- Location and Structure: PAM is typically found 3 to 4 nucleotides downstream of the nuclease cleavage site. Contrary to some misconceptions, the PAM sequence is not part of the bacterial immune system. Instead, it is present on the target phage or plasmid. Notably, the spacer within the bacterial CRISPR does not contain PAM.
- Function in Spacer Acquisition: The acquisition of spacers, a crucial step in the CRISPR mechanism, involves the integration of foreign DNA into the CRISPR locus. PAM plays a pivotal role in this process. Cas proteins, along with other components like the leader sequence and repeat region, recognize PAM for accurate spacer integration. If PAM is unidentifiable or lost due to mutations, the acquisition process is halted.
- Role in Interference: The interference mechanism in bacteria aims to degrade foreign DNA. While some studies suggest that PAM doesn’t play a significant role in interference, others indicate that the absence or mutation of specific PAM sequences can disrupt the interference mechanism. The exact role of PAM in interference remains a topic of ongoing research.
- Significance in Gene Editing: In the context of gene editing, PAM enhances the precision and accuracy of the CRISPR-Cas system. While the presence of PAM can sometimes pose challenges, especially if the target gene lacks a PAM sequence, it adds an extra layer of specificity to the editing process. Researchers often need to identify the PAM sequence and match it with the appropriate Cas protein for effective gene editing.
- Recent Advancements: Innovations in the field have led to the development of PAMless systems that can target entire genomes rather than specific loci. Such systems, like the one described by Walton et al., 2020, eliminate the need for PAM, offering a broader range of target sites. However, the efficiency and accuracy of these systems are still under evaluation.
Noteworthy Points:
- PAM serves as a marker, guiding the Cas proteins to recognize and cleave the protospacer, facilitating its integration into the CRISPR system.
- PAM is essential for DNA cleavage by Cas proteins but is not involved in RNA cleavage.
- The presence or absence of PAM can influence the efficiency of the CRISPR system, emphasizing its significance in genetic engineering.
In conclusion, the PAM sequence is an integral component of the CRISPR system, influencing its efficiency and accuracy. Its role in spacer acquisition, interference, and gene editing underscores its importance in the realm of genetic research and applications.
Application of CRISPR-CAS9
CRISPR gene editing
- In the food and agriculture industries, CRISPR has been used to engineer probiotic cultures and immunise industrial cultures (such as yoghurt) against illnesses. It is also used to increase crop output, drought resistance, and nutritional value.
- By the end of 2014, approximately one thousand research papers mentioning CRISPR had been published.
- The approach had been used to functionally inactivate genes in human cell lines and cells, to research Candida albicans, to genetically edit agricultural strains, and to modify yeasts used to produce biofuels.
- According to Hsu and his colleagues, the capacity to adjust genetic sequences enables reverse engineering, which has a favourable impact on biofuel production. CRISPR can also be used to modify mosquitos so that they cannot spread diseases such as malaria.
- Recently, a large variety of plant species have been successfully modified using Cas12a-based CRISPR techniques.
- A genetic disease patient was experimentally treated with CRISPR in July 2019. A 34-year-old lady with sickle cell illness was the patient.
- Using LASER ART, a novel antiretroviral medicine, and CRISPR, sixty to eighty percent of the integrated viral DNA was eliminated from animals in February 2020, and some mice were fully free of the virus.
- In an effort to treat Leber congenital amaurosis, a patient’s eye was injected with a CRISPR-modified virus in March 2020.
- CRISPR gene editing could potentially be utilised in the future to create new species or resurrect extinct species from closely related populations.
- Reevaluations of gene-disease correlations using CRISPR have led to the discovery of potentially significant abnormalities.
CRISPR as diagnostic tool
- Due to their capacity to specifically target nucleic acid sequences against a high background of non-target sequences, CRISPR-associated nucleases have proven effective as a tool for molecular testing.
- In 2016, the Cas9 nuclease was utilised to remove undesirable nucleotide sequences from next-generation sequencing libraries with only 250 picograms of RNA input.
- In 2017, CRISPR-associated nucleases were also employed for direct diagnostic testing of nucleic acids, with sensitivity down to a single molecule. By Martins et al. (2019), CRISPR diversity is utilised as an analytic target to determine phylogeny and diversity in bacteria, such as xanthomonads.
- As established by Shen et al., 2020, early identification of plant infections by molecular typing of the pathogen’s CRISPRs can be utilised in agriculture.
- By integrating CRISPR-based diagnostics with additional enzymatic activities, it is possible to identify substances other than nucleic acids. SHERLOCK-based Profiling of IN vitro Transcription is one example of a linked technology (SPRINT). SPRINT can be used to identify a number of compounds, such as metabolites in patient samples or environmental toxins, with high throughput or with portable point-of-care devices.
- CRISPR/Cas platforms are also being investigated for the detection and elimination of SARS-CoV-2, the virus responsible for COVID-19.
- For SARS-CoV-2, the AIOD-CRISPR and SHERLOCK assays have been identified as thorough diagnostic options.
- The SHERLOCK test is based on a press reporter RNA that is fluorescently labelled and capable of identifying 10 copies per microliter. AIOD-CRISPR facilitates robust and sensitive optical detection of viral nucleic acid.
Limitations of CRISPR-Cas9 system
CRISPR/Cas is a highly effective technique, yet it has significant limitations. It is:
- It is difficult to transfer large quantities of CRISPR/Cas material to mature cells, which is a difficulty for many therapeutic applications. Viral vectors are the most prevalent method of transmission.
- not 100% effective, therefore even cells that take in CRISPR/Cas may not change their genomes.
- Not one hundred percent accurate, and “off-target” modifications, although uncommon, can have serious repercussions, especially in therapeutic applications.
Potential Risks And Ethical Concerns Of Using Crispr
- Numerous possible applications of CRISPR technology raise problems regarding the ethics and repercussions of genome modification.
- The 2018 announcement by He Jiankui, a former biophysicist at the Southern University of Science and Technology in Shenzhen, that his team had modified DNA in human embryos and therefore generated the world’s first gene-edited newborns sparked numerous ethical arguments.
- Live Science earlier reported that he was sentenced to three years in prison and fined 3 million yuan for practising medicine without a licence, violating Chinese restrictions on human-assisted reproductive technology, and forging ethical review documents. However, even after his punishment, He’s experiments sparked questions about how the future use of CRISPR should be governed, given the relative youth of the technology.
- Obviously, illegal research on human embryos is an extreme misuse of CRISPR, but scientists assert that even ostensibly legitimate applications of the technology contain risks.
- Germline editing is the process of applying genetic adjustments to human embryos and reproductive cells, such as sperm and eggs. As modifications to these cells can be passed on to future generations, the use of CRISPR technology to modify the germline has raised a variety of ethical concerns.
- Safety issues are posed by variable efficacy, off-target consequences, and inaccurate editing. In addition, there is still much that the scientific community does not understand. In a 2015 article published in Science, David Baltimore and a group of scientists, ethicists, and legal experts note that germline editing may have unintended consequences for future generations “due to our limited understanding of human genetics, gene-environment interactions, and the pathways of disease” (including the interplay between diseases or conditions in the same patient).
- In a 2014 study published in Science, Oye and colleagues discuss the potential ecological effects of gene drives. Through crossbreeding, an introduced trait could extend beyond the intended population to other creatures. Additionally, gene drives could limit the genetic variety of the target population, which could hinder its ability to thrive.
- Other ethical issues are complicated in nature. Should we make significant changes that could affect future generations without their consent? What if germline editing evolves from a therapeutic tool to a tool for enhancing various human characteristics?
- To address these concerns, the National Academies of Sciences, Engineering, and Medicine have compiled a detailed report with recommendations and guidelines for genome editing.
- The National Academies advocate caution in the pursuit of germline editing, but they underscore that “caution does not mean prohibition.” They recommend that germline editing be performed only on genes that contribute to significant disorders and only when there are no other viable therapy options. They emphasise, among other things, the need to collect information on the risks and benefits to health and to maintain constant oversight during clinical trials. In addition, they urge that, following the conclusion of a trial, trial organisers follow up with the participants’ relatives for numerous generations to see which alterations in the genome remain over time.
Who Discovered Crispr?
- According to Quanta Magazine, scientists initially identified CRISPRs in bacteria in 1987, but they did not comprehend the biological importance of the DNA sequences and did not yet label them “CRISPRs” (opens in new tab).
- Yoshizumi Ishino and his colleagues at Osaka University in Japan discovered the distinctive nucleotide repeats and spacers in the intestinal bacteria Escherichia coli. As genetic analysis technology advanced in the 1990s, other researchers discovered CRISPRs in a variety of other microbes.
- According to a 2016 report(opens in new tab) in the journal Cell, Francisco Mojica, a researcher at the University of Alicante in Spain, was the first to characterise the unique characteristics of CRISPRs and discover the sequences in 20 different bacteria. Initially, he referred to the sequences as “short regularly spaced repeats” (SRSRs), but later he proposed that they be referred to as CRISPRs instead. Mojica has been in touch with Ruud Jansen of Utrecht University, who coined the name CRISPR in a 2002 article published in the journal Molecular Microbiology.
- In the years that followed, scientists also found Cas genes and the function of Cas enzymes, as well as the origin of the spacers in CRISPRs, as reported by Quanta.
- Jennifer Doudna, a professor of biochemistry, biophysics, and structural biology at the University of California, Berkeley, and Emmanuelle Charpentier, the director of the Max Planck Unit for the Science of Pathogens, shared the Nobel Prize in chemistry in 2020. Previously reported by Live Science, the two scientists are credited with turning the bacterial CRISPR/Cas system into a useful gene-editing tool.
- Charpentier discovered tracrRNA while studying the bacteria Streptococcus pyogenes, which is responsible for a variety of illnesses ranging from tonsillitis to sepsis. Charpentier began collaborating with Doudna to construct the CRISPR/Cas system when tracrRNA was identified as a previously unknown component. In 2012, the group announced in the journal Science that they had effectively transformed the molecular scissors into a gene-editing tool in their seminal work.
- Science Magazine noted that others believed Feng Zhang, a scientist at the Broad Institute, may also win the Nobel Prize for his independent contributions to the CRISPR system. Zhang revealed that the CRISPR system functions in mammalian cells; thus, the Broad Institute was awarded the first patent for the use of CRISPR gene-editing technology in eukaryotes, or complex cells with nuclei that contain DNA.
How Crispr Works As A Genome-Editing Tool described in Summary?
- Genomes encode a set of messages and instructions inside their DNA sequences. Genome editing entails modifying these DNA sequences, hence altering the encoded messages. This can be accomplished by placing a cut or break in the DNA and deceiving the cell’s normal DNA repair machinery into making the desired modifications. CRISPR-Cas9 provides a mechanism to do so.
- In 2012, two major scientific articles were published in the journals Science and PNAS that described how the bacterial CRISPR-Cas9 could be used to cut up any DNA, not simply that of viruses. Thus, the natural CRISPR system might be converted into a straightforward, programmable genome-editing instrument.
- Scientists may simply alter the sequence of the crRNA, which binds to a complementary sequence in the target DNA, the research concluded.In the 2012 Science publication, Martin Jinek and his colleagues further simplified the technique by merging crRNA and tracrRNA to create a single “guide RNA.” Thus, genome editing requires only two components: a guide RNA and the Cas9 protein.
- George Church, a professor of genetics at Harvard Medical School, told Live Science, “Operationally, you create a stretch of 20 base pairs that match a gene that you wish to alter.” From there, one may determine the complementary crRNA sequence. Church underlined the need of ensuring that the nucleotide sequence is unique to the target gene and not present anywhere else in the genome.
- Church added, “then the RNA plus the protein [Cas9] will cut, like a pair of scissors, the DNA at that place, and ideally nowhere else.” Once the DNA is cut, the cell’s inherent repair mechanisms kick in and begin to reassemble the DNA; at this stage, genome modifications are possible. There are two possible outcomes:
- According to the Stanford University Huntington’s Outreach Project, one form of mending involves glueing the two cuts back together. This technique, termed as “non-homologous end joining,” has a tendency to introduce errors in which nucleotides are inadvertently added or deleted, resulting in mutations that may disrupt a gene.
- In the second procedure, the break is repaired by inserting a nucleotide sequence into the gap. To accomplish this, the cell employs a short DNA strand as a template. Scientists can provide the DNA template of their choosing, allowing them to insert any gene or fix a mutation.
References
- Almendros C, Guzmán NM, Díez-Villaseñor C, García-Martínez J, Mojica FJ. Target motifs affecting natural immunity by a constitutive CRISPR-Cas system in Escherichia coli. PLoS One. 2012;7(11):e50797. doi: 10.1371/journal.pone.0050797. Epub 2012 Nov 26. PMID: 23189210; PMCID: PMC3506596.
- Lopez-Sanchez MJ, Sauvage E, Da Cunha V, Clermont D, Ratsima Hariniaina E, Gonzalez-Zorn B, Poyart C, Rosinski-Chupin I, Glaser P. The highly dynamic CRISPR1 system of Streptococcus agalactiae controls the diversity of its mobilome. Mol Microbiol. 2012 Sep;85(6):1057-71. doi: 10.1111/j.1365-2958.2012.08172.x. Epub 2012 Jul 27. PMID: 22834929.
- Shah SA, Hansen NR, Garrett RA. Distribution of CRISPR spacer matches in viruses and plasmids of crenarchaeal acidothermophiles and implications for their inhibitory mechanism. Biochem Soc Trans. 2009 Feb;37(Pt 1):23-8. doi: 10.1042/BST0370023. PMID: 19143596.
- Lillestøl RK, Shah SA, Brügger K, Redder P, Phan H, Christiansen J, Garrett RA. CRISPR families of the crenarchaeal genus Sulfolobus: bidirectional transcription and dynamic properties. Mol Microbiol. 2009 Apr;72(1):259-72. doi: 10.1111/j.1365-2958.2009.06641.x. Epub 2009 Feb 23. PMID: 19239620.
- Garcia-Heredia I, Martin-Cuadrado AB, Mojica FJ, et al. Reconstructing viral genomes from the environment using fosmid clones: the case of haloviruses. PLoS One. 2012;7(3):e33802. doi:10.1371/journal.pone.0033802.
- SIMPSON, B., DAVIDSON, B., & MAS MONTEYS, A. (2020, October 29). CRISPR/Cas9 gene editing of ATXN2 for the treatment of spinocerebellar ataxia type 2. WO2020219870 CRISPR/CAS9 GENE EDITING OF ATXN2 FOR THE TREATMENT OF SPINOCEREBELLAR ATAXIA TYPE 2.
- Ashizawa T, Öz G, Paulson HL. Spinocerebellar ataxias: prospects and challenges for therapy development [published correction appears in Nat Rev Neurol. 2018 Dec;14(12):749]. Nat Rev Neurol. 2018;14(10):590-605. doi:10.1038/s41582-018-0051-6.
- He L, Wang S, Peng L, et al. CRISPR/Cas9 mediated gene correction ameliorates abnormal phenotypes in spinocerebellar ataxia type 3 patient-derived induced pluripotent stem cells. Transl Psychiatry. 2021;11(1):479. Published 2021 Sep 17. doi:10.1038/s41398-021-01605-2.
- Salvatori F & Pappada M et al., (2020). “CRISPR/CAS9-based silencing of the ATXN1 gene in Spinocerebellar ataxia type I (SCAI) fibroblast. bioRxiv- Cold Spring Harbor Laboratory.
- Ran, F., Hsu, P., Wright, J. et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8, 2281–2308 (2013). https://doi.org/10.1038/nprot.2013.143
- https://www.yourgenome.org/facts/what-is-crispr-cas9/
- https://crisprtx.com/gene-editing/crispr-cas9
- https://www.jax.org/personalized-medicine/precision-medicine-and-you/what-is-crispr
- https:/en.wikipedia.org/wiki/Genome_editing
- https://geneticeducation.co.in/what-is-gene-editing-and-crispr-cas9/
- https://www.livescience.com/58790-crispr-explained.html
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.