What is Cloning Vector?
A cloning vector is a small portion of DNA that can be maintained in an organism and into which a foreign DNA fragment can be inserted for the purposes of cloning. The vector for cloning may consist of DNA extracted from a virus, the cell of a higher organism, or the plasmid of a bacterium. The vector includes characteristics that facilitate the insertion or removal of a DNA fragment, such as the presence of restriction sites. Vector DNA and foreign DNA can be treated with a restriction enzyme that cuts the DNA to generate DNA fragments with blunt or adhesive ends. Vector DNA and foreign DNA with compatible ends can then be joined via molecular ligation. After a DNA fragment has been cloned into a cloning vector, it may be subcloned into a vector with a more specific application.
There are numerous types of cloning vectors, but genetically modified plasmids are the most common. Plasmids, bacteriophages (such as phage ), cosmids, and bacterial artificial chromosomes (BACs) are examples of cloning vectors in Escherichia coli. Some DNA, such as very large DNA fragments, cannot be maintained in E. coli, so other organisms, such as yeast, must be used. Yeast vectors for cloning contain yeast artificial chromosomes (YACs).
Purpose of cloning vector
The purpose of a cloning vector is to facilitate the replication and manipulation of DNA fragments in a host organism. Cloning vectors serve as vehicles for the insertion and propagation of foreign DNA fragments, allowing researchers to amplify, study, and modify specific DNA sequences of interest. The key purposes of cloning vectors include:
- Gene Cloning: Cloning vectors enable the isolation and amplification of specific genes or DNA fragments for further analysis, such as gene sequencing, functional characterization, or gene expression studies.
- DNA Manipulation: Cloning vectors provide a platform for DNA manipulation techniques, including the insertion, deletion, or mutation of specific DNA sequences. These manipulations help elucidate gene function, regulatory elements, or protein structure and function.
- DNA Libraries: Cloning vectors are used to construct DNA libraries, which are collections of cloned DNA fragments that represent an organism’s entire genome or a specific subset of genes. DNA libraries serve as valuable resources for genomic research, gene discovery, and screening for specific DNA sequences of interest.
- Protein Production: Certain cloning vectors, such as expression vectors, are designed for the efficient expression of cloned genes in host organisms. These vectors enable the production of recombinant proteins for various applications, including research, biotechnology, and therapeutic purposes.
- Genetic Engineering: Cloning vectors are essential tools in genetic engineering for the creation of genetically modified organisms (GMOs) or the generation of transgenic organisms. They facilitate the insertion of foreign DNA into the genome of the host organism, allowing the introduction of desired traits or the study of gene function.
- Functional Analysis: Cloning vectors are used to investigate gene function by introducing cloned genes into host organisms and studying the resulting phenotypic changes. This allows researchers to determine the role of specific genes in biological processes or disease mechanisms.
- DNA Sequencing: Cloning vectors play a crucial role in DNA sequencing methods, such as Sanger sequencing or next-generation sequencing (NGS). Cloning of DNA fragments into vectors allows for the amplification and isolation of the target DNA, which can then be sequenced to determine its nucleotide sequence.
- Gene Therapy: Cloning vectors, particularly viral vectors, are utilized in gene therapy approaches to deliver therapeutic genes into target cells or tissues. These vectors help in the treatment of genetic disorders, cancer, and other diseases by replacing defective or missing genes with functional ones.
How are cloning vectors different from other types of vectors?
Cloning vectors are distinct from other types of vectors based on their specific features and intended purposes. Here are some key differences between cloning vectors and other commonly used vectors:
- Cloning Purpose: The primary purpose of cloning vectors is to facilitate the replication and manipulation of DNA fragments. They are designed to carry and propagate foreign DNA sequences in a host organism. Other types of vectors, such as expression vectors or viral vectors, have additional features tailored for specific purposes, such as gene expression or gene delivery.
- Insert Size: Cloning vectors typically have a larger capacity to accommodate DNA inserts compared to other vectors. They can carry DNA fragments ranging from a few hundred base pairs to several kilobases, allowing for the cloning of genes or genomic regions of interest.
- Selectable Markers: Cloning vectors commonly incorporate selectable markers, such as antibiotic resistance genes, which enable the selection and identification of host cells that have successfully taken up and propagated the vector. These markers facilitate the isolation of cells with the desired cloning construct. In contrast, other vectors may use different types of selectable markers specific to their intended application, such as reporter genes for expression vectors or viral genes for viral vectors.
- Expression Elements: Cloning vectors typically do not contain strong promoter elements or other expression-related features. Their main purpose is to clone DNA fragments, and the expression of the inserted genes is often limited or not a primary focus. In contrast, expression vectors are specifically designed for gene expression, incorporating strong promoters, enhancers, translation initiation sequences, and other elements to facilitate high-level protein production.
- Host Compatibility: Cloning vectors are designed to be compatible with a wide range of host organisms, including bacteria, yeast, and other organisms used in molecular biology research. They are engineered to function effectively in these hosts for DNA replication and propagation. In contrast, other vectors may be tailored for specific host organisms or cell types, such as viral vectors for gene delivery into mammalian cells or specific vectors for insect or plant systems.
- Copy Number: Cloning vectors often have a higher copy number within the host cells, allowing for efficient amplification of the cloned DNA. This is advantageous for obtaining sufficient amounts of the cloned DNA for downstream applications. Other types of vectors may have lower copy numbers or regulated replication mechanisms, depending on their specific purpose and host system.
- Additional Features: Cloning vectors typically lack additional features like viral replication origins, viral envelope proteins, or specific sequences required for packaging into viral particles. These features are present in viral vectors, which are designed for efficient gene delivery and expression in target cells.
Types of selectable markers used in cloning vectors
Cloning vectors often incorporate selectable markers, which are genes or genetic elements that confer a selective advantage to host cells that have successfully taken up and maintained the vector. Selectable markers allow researchers to identify and isolate cells that carry the desired cloning construct. Here are some common types of selectable markers used in cloning vectors:
- Antibiotic Resistance Genes: Antibiotic resistance genes are widely used as selectable markers in cloning vectors. These genes provide resistance to specific antibiotics, allowing the growth and survival of host cells that have taken up the vector in the presence of the corresponding antibiotic. Examples include ampicillin resistance (ampR), kanamycin resistance (kanR), chloramphenicol resistance (cat), and tetracycline resistance (tetR).
- Herbicide Resistance Genes: In plant molecular biology research, herbicide resistance genes are used as selectable markers. These genes encode enzymes that confer resistance to specific herbicides, allowing the growth and survival of transgenic plants in the presence of the herbicide. Common examples include glyphosate resistance (e.g., EPSPS gene) and glufosinate resistance (e.g., BAR gene).
- Auxotrophic Markers: Auxotrophic markers are used as selectable markers in yeast cloning vectors. These markers are based on mutations in genes involved in essential biosynthetic pathways, rendering the host yeast strain unable to synthesize certain nutrients. By providing the missing nutrient in the growth medium, only yeast cells that have taken up the vector and acquired the functional auxotrophic marker gene can survive.
- Reporter Genes: Reporter genes are selectable markers commonly used in expression vectors to study gene expression or protein localization. These genes encode enzymes or fluorescent proteins that produce detectable signals, allowing researchers to easily visualize or quantify the expression of the gene of interest. Examples include β-galactosidase (lacZ), green fluorescent protein (GFP), and luciferase (luc).
- Genetic Complementation Markers: Genetic complementation markers are used when host cells carry a mutation in a specific gene. The cloning vector contains a functional copy of the mutated gene, allowing the complementation of the mutant phenotype. This allows for the selection and identification of host cells that have taken up the vector based on their restored functionality.
Are there different types of promoters available in cloning vectors?
Yes, there are different types of promoters available in cloning vectors. Promoters are DNA sequences located upstream of the gene of interest that regulate its transcription and determine the level and specificity of gene expression. In cloning vectors, the choice of promoter depends on the desired level and regulation of gene expression in the host organism. Here are some common types of promoters used in cloning vectors:
- Constitutive Promoters: Constitutive promoters are active under normal growth conditions and drive continuous, high-level expression of the gene of interest. They provide stable and consistent expression throughout various stages of growth and in different cell types. Examples include the commonly used promoters such as the cytomegalovirus (CMV) promoter, the human elongation factor 1 alpha (EF1α) promoter, and the SV40 early promoter.
- Tissue-Specific Promoters: Tissue-specific promoters are active only in specific cell types or tissues, allowing targeted expression of the gene of interest. These promoters can be derived from genes that are known to be specifically expressed in certain cell types or tissues. By using tissue-specific promoters, researchers can achieve gene expression that is restricted to specific cell populations. Examples include the muscle-specific promoter (e.g., muscle creatine kinase promoter) or neuron-specific promoter (e.g., synapsin promoter).
- Inducible Promoters: Inducible promoters allow researchers to control the timing and level of gene expression by responding to specific inducers or environmental cues. These promoters are inactive or have low basal activity in the absence of the inducer and can be activated or repressed by adding or removing specific substances. Examples of inducible promoters include the tetracycline-inducible (Tet-On/Tet-Off) promoter system and the ecdysone-inducible promoter system.
- Promoters with Enhancer Elements: Promoters can be combined with enhancer elements to increase the level of gene expression. Enhancers are DNA sequences that enhance the activity of promoters and can be located either upstream or downstream of the promoter region. The presence of enhancer elements can lead to stronger and more robust gene expression. Common examples include the CMV enhancer-promoter combination.
- Cell-Type-Specific Promoters: In some cases, promoters specific to a particular cell type or lineage are used to drive gene expression in a specific cell population. These promoters are often derived from genes that are highly expressed in the desired cell type or have important roles in its development or function. Cell-type-specific promoters are commonly used in developmental biology and stem cell research.
Characteristics of a cloning vector
- Origin of Replication: Cloning vectors contain an origin of replication (ori), which is a DNA sequence recognized by the host organism’s replication machinery. It allows the vector to replicate autonomously within the host cells.
- Selectable Marker: Cloning vectors carry selectable markers, such as antibiotic resistance genes or genes for nutrient utilization, which allow for the selection of host cells that have taken up the vector during transformation. Only cells containing the vector and the inserted DNA fragment survive in the presence of the selective agent.
- Multiple Cloning Site (MCS): The MCS, also known as a polylinker or cloning site, is a short DNA region within the cloning vector that contains multiple unique restriction enzyme recognition sites. It enables convenient insertion of foreign DNA fragments at specific locations.
- Promoters: Cloning vectors often possess promoter sequences that drive the transcription of inserted DNA fragments in the host organism. These promoters can be inducible or constitutive, allowing control over gene expression.
- Terminator Sequences: Terminator sequences are present in cloning vectors to ensure proper termination of transcription of the inserted DNA fragment. They provide signals for the transcription machinery to stop transcription accurately.
- Plasmid Backbone: Cloning vectors are typically plasmids, which are circular DNA molecules. The plasmid backbone contains essential elements, including replication origins, selectable markers, and regulatory elements, that maintain the stability and propagation of the vector in the host cells.
- Size and Copy Number: Cloning vectors vary in size and copy number. The size of the vector affects the capacity for accommodating large DNA inserts, while the copy number influences the abundance of the vector within the host cells.
- Sequencing and Reporter Tags: Some cloning vectors include specific sequences or tags that facilitate DNA sequencing or enable the detection and quantification of the inserted DNA fragment, such as fluorescent or enzymatic reporter genes.
- Compatibility with Different Hosts: Cloning vectors are designed to be compatible with specific host organisms, such as bacteria (e.g., Escherichia coli), yeast (e.g., Saccharomyces cerevisiae), or other cell types. The vector components and features must be compatible with the host’s replication and gene expression machinery.
- DNA Purification Tags: Certain cloning vectors include tags or markers that facilitate purification of the recombinant DNA, allowing for easy isolation of the inserted DNA fragment using affinity chromatography or other purification techniques.

| Feature | Description |
|---|---|
| Origin of Replication | Allows autonomous replication of the vector within host cells |
| Selectable Marker | Enables selection of transformed cells through antibiotic resistance or nutrient utilization |
| Multiple Cloning Site | Contains unique restriction enzyme recognition sites for easy insertion of DNA fragments |
| Promoters | Drives transcription of inserted DNA fragments in the host organism |
| Terminator Sequences | Ensures proper termination of transcription of the inserted DNA fragment |
| Plasmid Backbone | Provides essential elements for vector stability and propagation |
| Size and Copy Number | Varies in size, affecting the capacity for large DNA inserts, and copy number influences abundance in host cells |
| Sequencing and Reporter Tags | Facilitates DNA sequencing and enables detection or quantification of the inserted DNA fragment |
| Compatibility with Different Hosts | Designed for specific host organisms and their replication and gene expression machinery |
| DNA Purification Tags | Allows for easy isolation of the inserted DNA fragment during purification |
Origin of replication (Ori) in the host cell
The origin of replication (Ori) is a specific DNA sequence within the genome of a host cell where the replication process initiates. It is crucial for the replication of DNA molecules, including cloning vectors, within the host cell. The Ori sequence serves as a recognition site for the replication machinery of the host cell, allowing it to bind and initiate the replication process.
When a cloning vector carrying an Ori sequence is introduced into a host cell, the host’s replication machinery recognizes the Ori and initiates the replication of the vector DNA. This results in the amplification and propagation of the vector within the host cell. The replicated vector DNA can then be passed on to daughter cells during cell division, ensuring the maintenance and stability of the vector within a population of cells.
The Ori sequence is specific to the host organism and is designed to be compatible with the replication machinery of that organism. Different host organisms, such as bacteria or yeast, have distinct Ori sequences that are recognized by their respective replication enzymes.
By utilizing the host cell’s own replication machinery, the presence of an Ori sequence in a cloning vector allows for the autonomous replication of the vector DNA, independent of the host’s genomic DNA replication. This feature enables the production of multiple copies of the vector, which is advantageous for various applications in molecular biology, such as amplifying and studying specific DNA fragments or producing large quantities of recombinant proteins.
Significance of origin of replication (ori)
- Autonomous Replication: The presence of an ori in a cloning vector allows the vector to replicate independently within the host cell. It ensures that the inserted DNA fragment, along with the vector, is faithfully replicated and maintained during cell division. This autonomy enables the generation of multiple copies of the vector and the inserted DNA, facilitating the amplification of the desired DNA sequence.
- Propagation of Cloned DNA: The ori sequence ensures the propagation and stability of the cloning vector in host cells over multiple generations. As the host cell divides, the replicated vector DNA is passed on to daughter cells, preserving the inserted DNA fragment. This propagation enables the maintenance of the cloned DNA for downstream applications and further genetic analysis.
- Generation of Sufficient DNA: The ori’s role in autonomous replication allows for the production of an abundant quantity of the cloned DNA. This feature is particularly valuable when large amounts of the DNA fragment are required for subsequent experiments, such as sequencing, protein expression, or functional analysis.
- DNA Manipulation and Engineering: The presence of ori facilitates the cloning and manipulation of DNA fragments. Researchers can introduce specific DNA sequences into a cloning vector containing an ori, allowing the generation of recombinant DNA molecules. The ability to manipulate and engineer DNA is essential for a range of applications, including gene cloning, gene expression analysis, and genetic engineering.
- Efficient Screening and Selection: Cloning vectors carrying an ori often include selectable markers, such as antibiotic resistance genes. These markers enable the selection and identification of host cells that have successfully taken up the cloning vector. The ori ensures that the selected cells, containing both the vector and the inserted DNA, can be efficiently replicated and maintained.
- Versatility across Host Organisms: Different host organisms have specific ori sequences that are recognized by their respective replication machinery. Cloning vectors can be designed with compatible ori sequences for various host organisms, such as bacteria, yeast, or mammalian cells. This versatility allows researchers to choose the appropriate vector and host system for their specific cloning needs.
Why are vectors with a stringent control ori low copy number?
Vectors with a stringent control origin of replication (ori) are designed to have a low copy number in host cells. The low copy number is intentional and serves several important purposes in molecular cloning. Here are the reasons why vectors with a stringent control ori are typically low copy number:
- Minimize Cellular Burden: High copy number vectors can place a significant burden on the host cell’s resources and cellular machinery. The replication and maintenance of multiple copies of a high copy number vector consume a substantial amount of energy and cellular components. By using a low copy number vector, the strain on the host cell is reduced, allowing it to allocate its resources more efficiently for other essential cellular processes.
- Stable Maintenance: Low copy number vectors tend to exhibit more stable maintenance within host cells. With fewer copies of the vector present, the chances of segregation errors during cell division or loss of the vector due to genetic instability are reduced. This stability ensures the long-term maintenance and preservation of the vector and the inserted DNA fragment.
- Minimize Toxic Effects: Some high copy number vectors can induce toxic effects on the host cells. The overproduction of certain proteins or RNA molecules encoded by high copy number vectors can lead to cellular stress, growth inhibition, or even cell death. By using low copy number vectors, the expression levels of potentially toxic components are kept at a manageable level, minimizing adverse effects on the host cells.
- Maintain Plasmid Integrity: High copy number vectors are more prone to rearrangements, deletions, or mutations due to increased homologous recombination events or replication errors. Low copy number vectors experience fewer replication and recombination events, reducing the chances of genetic alterations and maintaining the integrity of the plasmid and its inserted DNA fragment.
- Reduce Background Noise: In certain experiments, such as gene expression analysis or protein production, it is desirable to have a low background noise level. High copy number vectors can result in increased background noise due to unintended transcription, translation, or non-specific interactions. Low copy number vectors help minimize this background noise and enhance the sensitivity and specificity of the desired experimental outcomes.
Mechanism of relaxed control of vector replication
The relaxed control of vector replication refers to the ability of certain vectors to undergo replication in a less stringent manner, leading to an increased copy number within host cells. Unlike vectors with stringent control origins of replication (ori) that maintain low copy numbers, vectors with relaxed control ori can replicate more frequently and generate higher numbers of vector copies. The mechanism of relaxed control of vector replication typically involves the following aspects:
- Altered Origin of Replication: Vectors with relaxed control of replication often possess modified or mutated origins of replication. These modifications can affect the binding affinity of replication initiation proteins or alter the interaction between the origin and the replication machinery. These changes lead to a relaxed or loosened control over the replication process, allowing for more frequent initiation events.
- Enhanced Binding of Replication Proteins: Relaxed control vectors may have mutated or modified sequences within the ori that increase the binding affinity of replication initiation proteins. These proteins play a crucial role in the initiation of DNA replication by recognizing and binding to the ori. With enhanced binding affinity, replication proteins are more efficiently recruited to the ori, promoting frequent replication initiation events.
- Reduced Negative Regulation: In some cases, the relaxed control of vector replication involves mutations or modifications that decrease the negative regulation of replication. Negative regulatory elements or proteins normally act to suppress or limit replication initiation. Alterations in these elements can reduce their inhibitory effects, allowing for a higher frequency of replication initiation events.
- Increased Efficiency of Replication Machinery: Relaxed control vectors may also exhibit modifications that enhance the efficiency or processivity of the replication machinery. These modifications can improve the speed or accuracy of DNA synthesis during replication, leading to more rapid and efficient replication cycles. The increased efficiency contributes to the overall higher copy number of the vector within host cells.
- Host-Specific Factors: The relaxed control of replication can also be influenced by host-specific factors. Some host cells may possess endogenous factors or mechanisms that favor a more permissive replication environment. These factors can interact with the vector and its replication machinery to facilitate relaxed control of replication and increase the copy number.
Multiple Cloning Site (MCS)
The Multiple Cloning Site (MCS), also known as a polylinker or cloning site, is a short DNA sequence within a cloning vector that contains multiple unique restriction enzyme recognition sites. The MCS is a fundamental component of cloning vectors and serves as a convenient and versatile tool for the insertion of foreign DNA fragments into the vector. Here are some key features and functions of the Multiple Cloning Site:
- Multiple Restriction Enzyme Sites: The MCS contains several different recognition sites for various restriction enzymes. These sites are typically short DNA sequences that are specifically recognized and cleaved by restriction enzymes, which are enzymes that cut DNA at specific sequences. Each recognition site within the MCS is unique, allowing for the selective insertion of DNA fragments at specific locations within the vector.
- DNA Fragment Insertion: The primary function of the MCS is to facilitate the insertion of foreign DNA fragments into the cloning vector. Researchers can choose a suitable restriction enzyme to digest both the vector and the DNA fragment of interest. The compatible cohesive ends or overhangs generated by the restriction enzyme digestion in both the vector and the DNA fragment allow for their precise ligation. This results in the incorporation of the DNA fragment into the vector at the desired location within the MCS.
- Flexibility and Versatility: The presence of multiple unique restriction enzyme recognition sites within the MCS provides flexibility and versatility in the choice of insertion sites for DNA fragments. Researchers can select the appropriate restriction enzyme and corresponding recognition site based on factors such as the size of the DNA fragment, downstream applications, or specific experimental requirements.
- Orientation Control: The arrangement of the recognition sites within the MCS determines the orientation of the inserted DNA fragment within the vector. The order and spacing of the restriction enzyme sites in the MCS allow researchers to control the orientation of the inserted DNA fragment, which can be critical for downstream applications that require specific gene expression orientations or promoter-probe studies.
- Sequence Verification: The MCS often includes additional unique sequence features or short DNA sequences flanking the restriction enzyme recognition sites. These additional sequences aid in the verification and confirmation of successful cloning events. They can be used as primer binding sites for sequencing or as probes for specific diagnostic tests to ensure the correct insertion and orientation of the DNA fragment within the vector.
- Compatibility with Other Vectors: The MCS is designed to be compatible with various other cloning vectors and DNA manipulation techniques. The sequences of the restriction enzyme recognition sites within the MCS are carefully selected to avoid common recognition sites used in other cloning vectors. This compatibility allows for the transfer and exchange of DNA fragments between different vectors and experimental systems.
The Multiple Cloning Site is a crucial feature of cloning vectors, enabling the precise and efficient insertion of DNA fragments into the vector at specific locations. Its versatility, flexibility, and compatibility make the MCS an essential tool for a wide range of molecular biology applications, including gene cloning, recombinant DNA technology, and genetic engineering.
Selectable marker
A selectable marker, in the context of molecular cloning, refers to a gene or DNA sequence incorporated into a cloning vector that confers a selectable phenotype to host cells. The selectable marker allows researchers to selectively identify and isolate host cells that have successfully taken up and maintained the cloning vector during transformation or transfection experiments. Here are some key features and functions of selectable markers:
- Positive Selection: Selectable markers provide a means of positively selecting for host cells that have successfully incorporated the cloning vector. The selectable marker gene is usually linked to the DNA fragment of interest within the vector. When host cells are transformed or transfected with the vector, only cells that have taken up and retained the vector will express the selectable marker gene, providing a visible or measurable phenotype.
- Antibiotic Resistance: One of the most commonly used types of selectable markers is antibiotic resistance genes. These genes encode proteins that confer resistance to specific antibiotics. The host cells that have successfully taken up the vector with the selectable marker gene will be able to grow and survive in the presence of the corresponding antibiotic, while non-transformed or non-transfected cells will be inhibited or killed by the antibiotic.
- Nutrient Utilization: In addition to antibiotic resistance markers, selectable markers can also include genes that allow host cells to utilize specific nutrients that are otherwise non-utilizable by the wild-type cells. For example, certain marker genes enable host cells to metabolize or grow on unique carbon sources, amino acids, or other nutrient types. Only cells that have acquired and maintained the vector with the selectable marker gene can grow or survive under specific nutrient conditions.
- Reporter Genes: Selectable markers can also be designed as reporter genes, which produce a visible or detectable phenotype when expressed. These reporter genes encode enzymes, fluorescent proteins, or other proteins that can be easily visualized or measured. The expression of the reporter gene indicates successful uptake and retention of the cloning vector by host cells.
- Genetic Linkage: Selectable markers are often genetically linked to the DNA fragment of interest within the cloning vector. This linkage ensures that cells containing the selectable marker gene also carry the desired DNA sequence. By selecting for the presence of the selectable marker, researchers can indirectly identify and isolate cells that have incorporated the DNA fragment of interest.
- Facilitate Screening and Analysis: Selectable markers greatly simplify the process of screening and analyzing transformed or transfected cells. They enable researchers to selectively grow and isolate cells that have successfully incorporated the vector and express the selectable marker gene. This facilitates downstream experiments such as DNA sequencing, protein expression analysis, or functional characterization of the cloned DNA fragment.
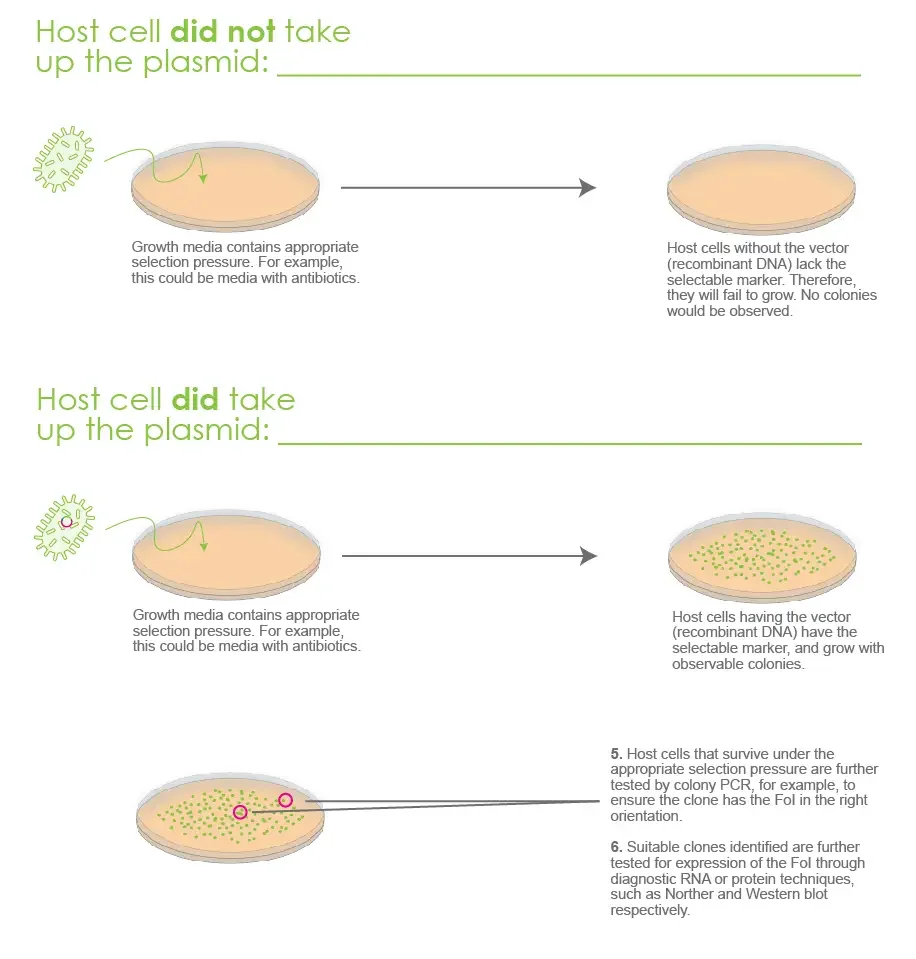
Selectable markers are essential components of cloning vectors, allowing for the identification, isolation, and analysis of host cells that have successfully taken up and maintained the vector. Their presence ensures efficient selection and enrichment of cells carrying the desired DNA fragment, streamlining the molecular cloning process.
Promoters
Promoters are DNA sequences that play a crucial role in initiating and regulating gene expression. They are located upstream of the coding sequence of a gene and serve as binding sites for transcription factors and RNA polymerase, which together initiate the transcription of the gene. Here are some key features and functions of promoters:
- Transcription Initiation: The primary function of promoters is to initiate the transcription of a gene. Promoters provide binding sites for RNA polymerase, the enzyme responsible for synthesizing RNA from a DNA template. When specific transcription factors and RNA polymerase bind to the promoter, they form a transcription initiation complex, which initiates the synthesis of an RNA molecule complementary to the gene’s coding sequence.
- Regulatory Elements: Promoters often contain additional regulatory elements such as enhancers, silencers, and response elements. These regulatory elements can be located near or within the promoter region and interact with specific transcription factors to modulate the rate of gene transcription. They can enhance or inhibit the binding of transcription factors and RNA polymerase to the promoter, resulting in the upregulation or downregulation of gene expression.
- TATA Box and Core Promoter Elements: Many promoters contain specific sequence motifs that are essential for efficient transcription initiation. One well-known element is the TATA box, a DNA sequence rich in adenine and thymine (AT) located around 25-30 base pairs upstream of the transcription start site. The TATA box is recognized by transcription factors and helps position RNA polymerase at the correct location for transcription initiation. Additionally, core promoter elements, such as the initiator (Inr) sequence, downstream promoter element (DPE), and downstream core element (DCE), can contribute to the efficiency and accuracy of transcription initiation.
- Specificity and Directionality: Promoters exhibit specificity and directionality in gene expression. Each gene has its own unique promoter sequence, allowing for the specific recognition and binding of transcription factors and RNA polymerase. The promoter sequence determines which genes are transcribed and in what direction. The orientation and structure of the promoter play a crucial role in determining which DNA strand is transcribed and which RNA strand is produced.
- Strength and Activity: Promoters can vary in their strength and activity levels, influencing the level of gene expression. Some promoters are constitutive, meaning they are active under normal conditions and drive consistent gene expression. Others are inducible or repressible, responding to specific signals or conditions to modulate gene expression accordingly. The strength and activity of a promoter are determined by its sequence elements, regulatory factors, and interactions with transcriptional activators or repressors.
- Cell-Type Specificity: Promoters can exhibit cell-type specificity, allowing for the regulation of gene expression in a cell- or tissue-specific manner. Certain promoters are active only in specific cell types or developmental stages, controlling the expression of genes that are required for specific cellular functions or processes. This cell-type specificity is achieved through the binding of cell-specific transcription factors and interactions with cell-specific regulatory elements.
Promoters are essential elements in gene expression regulation, controlling the initiation and level of transcription for specific genes. Their diversity, specificity, and interaction with regulatory factors contribute to the complex regulation of gene expression in different cell types, developmental stages, and environmental conditions.
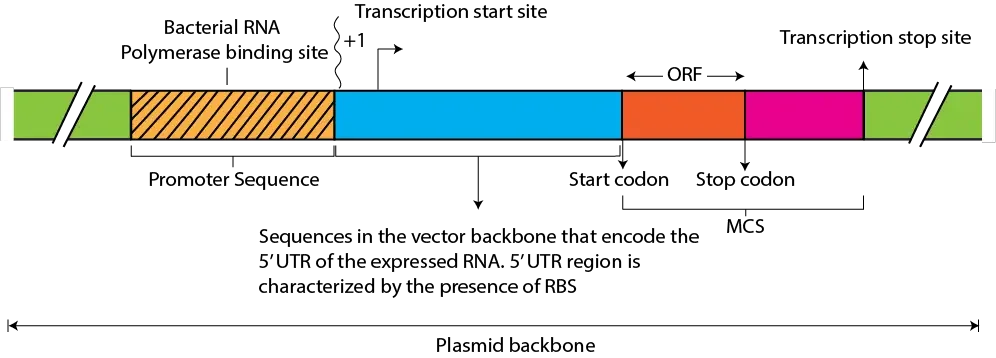
Terminator Sequences
Terminator sequences, also known as transcription terminators or polyadenylation signals, are DNA sequences that play a critical role in the termination of transcription in cells. They are typically located downstream of the coding sequence of a gene and are responsible for signaling the end of RNA synthesis. Here are some key features and functions of terminator sequences:
- Transcription Termination: The primary function of terminator sequences is to signal the termination of transcription. Once the RNA polymerase reaches a terminator sequence, it recognizes the specific DNA sequence and undergoes a series of structural changes. These changes lead to the release of the RNA transcript and detachment of the RNA polymerase from the DNA template, effectively terminating transcription.
- Structure and Elements: Terminator sequences often possess specific structural elements that facilitate the termination process. One common element is the presence of a termination hairpin or stem-loop structure formed by the folding of the RNA transcript. This structure is stabilized by complementary base pairing within the transcript, resulting in the dissociation of the RNA polymerase from the DNA template.
- Polyadenylation Signals: In eukaryotes, terminator sequences typically contain polyadenylation signals or poly(A) signals. These signals are specific DNA sequences that are recognized by proteins involved in the post-transcriptional processing of RNA. Polyadenylation signals direct the addition of a poly(A) tail to the 3′ end of the RNA transcript, which helps stabilize the transcript and influences its stability, transport, and translation efficiency.
- Efficiency of Termination: Terminator sequences can vary in their efficiency of transcription termination. Some terminators are more efficient, leading to a higher likelihood of complete termination and detachment of the RNA polymerase from the DNA template. Others may exhibit partial termination, resulting in read-through transcription and the production of longer RNA transcripts. The efficiency of termination can be influenced by various factors, including the sequence elements within the terminator and interactions with specific proteins or regulatory factors.
- Preventing Read-Through: Terminator sequences also serve to prevent read-through transcription, where RNA polymerase continues to transcribe beyond the intended termination site. The termination hairpin or stem-loop structure, along with associated factors, helps create a physical barrier for the progressing RNA polymerase, effectively stopping its movement and preventing read-through.
- Regulation of Gene Expression: Terminator sequences can contribute to the regulation of gene expression. By influencing the efficiency of transcription termination, terminators can affect the abundance and stability of RNA transcripts. Efficient termination ensures that the correct transcript length is produced, preventing the synthesis of aberrant or truncated transcripts. Additionally, the presence of specific terminator sequences can influence the choice of alternative polyadenylation sites, leading to the generation of different mRNA isoforms with varying stability and translational efficiency.
Terminator sequences are essential components of gene expression regulation, controlling the proper termination of transcription and the generation of functional RNA molecules. Their structure, sequence elements, and interactions with proteins or regulatory factors contribute to the accurate and efficient termination of transcription processes in cells.
Plasmid Backbone
A plasmid backbone refers to the DNA sequence or structural elements that are common to all plasmids of a particular type or class. It serves as the structural framework or scaffold for the insertion and maintenance of foreign DNA fragments in the plasmid. Here are some key features and functions of a plasmid backbone:
- Origin of Replication (Ori): The plasmid backbone contains an origin of replication (ori), which is a specific DNA sequence that is recognized and bound by replication proteins. The ori is responsible for initiating and controlling the replication of the plasmid within host cells. It ensures that the plasmid can replicate independently of the host chromosome.
- Selectable Marker: The plasmid backbone often includes a selectable marker gene, such as an antibiotic resistance gene. The selectable marker provides a means to select and identify host cells that have successfully taken up and maintained the plasmid. Cells carrying the plasmid can grow and survive in the presence of a specific antibiotic, while non-transformed or non-transfected cells are inhibited or killed by the antibiotic.
- Multiple Cloning Site (MCS): The plasmid backbone typically contains a multiple cloning site (MCS), also known as a polylinker. The MCS is a short DNA sequence that contains multiple unique restriction enzyme recognition sites. These sites allow for the insertion of foreign DNA fragments at specific locations within the plasmid. The MCS provides versatility and convenience in cloning experiments by offering a range of options for inserting DNA fragments.
- Promoters and Regulatory Elements: Plasmid backbones may contain promoters and regulatory elements that can drive the expression of genes or regulate the expression of inserted DNA fragments. These elements can include enhancers, silencers, response elements, or specific promoter sequences that allow for the controlled and regulated expression of genes or gene products.
- Replication Control Elements: Plasmid backbones often contain replication control elements that help regulate plasmid copy number. These elements include DNA sequences or proteins that can influence the initiation or termination of plasmid replication. By controlling the copy number, these elements ensure the appropriate balance between plasmid maintenance and stability within host cells.
- Size and Compatibility: Plasmid backbones can vary in size, depending on the specific plasmid type and the additional elements incorporated into the backbone. The size of the backbone can influence the efficiency of plasmid purification, transformation, and other molecular biology techniques. Plasmid backbones are also designed to be compatible with various host organisms, allowing for their replication and maintenance in specific bacterial, yeast, or mammalian cell systems.
The plasmid backbone provides the essential elements for replication, selection, cloning, and expression of foreign DNA fragments. It acts as a platform for the construction of recombinant plasmids and enables the study and manipulation of genes and gene products in molecular biology research.
Type of Cloning Vectors
There are several types of cloning vectors that are commonly used in molecular biology research. Each type of vector has specific features and characteristics that make it suitable for different cloning applications. Here are some of the main types of cloning vectors:
1. Plasmid Vectors
Plasmid vectors are circular DNA molecules that are commonly used for cloning small to medium-sized DNA fragments. They can replicate autonomously within bacterial cells and often contain features such as selectable markers, multiple cloning sites (MCS), and origin of replication (ori). Plasmids are versatile and widely used in molecular cloning experiments.
Here are some key features of plasmid vectors:
- Circular DNA Structure: Plasmid vectors have a circular structure, which allows for efficient replication and stability within bacterial cells. The circular DNA is resistant to degradation and provides a stable template for gene expression.
- Origin of Replication (Ori): Plasmid vectors contain an origin of replication (Ori) sequence that enables autonomous replication within host cells. The Ori sequence is recognized by the host cell machinery, initiating the replication process and ensuring the faithful duplication of the plasmid.
- Multiple Cloning Sites (MCS): Plasmid vectors have specific regions called multiple cloning sites or polylinker regions. These sites contain multiple unique restriction enzyme recognition sequences, allowing for easy insertion of DNA fragments of interest. The MCS simplifies the cloning process by providing a range of options for DNA fragment insertion.
- Selectable Markers: Plasmid vectors often carry selectable markers, such as antibiotic resistance genes. These markers allow for the identification and selection of host cells that have successfully taken up the plasmid. Cells without the plasmid or those that have lost it during replication will not survive in the presence of the selection agent.
- Copy Number: Plasmid vectors can exist in multiple copies within a bacterial cell. The copy number can vary depending on the specific plasmid and host system used. High-copy plasmids can exist in tens to hundreds of copies per cell, providing a high yield of plasmid DNA. Low-copy plasmids have fewer copies per cell, resulting in lower plasmid yield but reduced metabolic burden on the host cell.
- Promoters and Regulatory Elements: Plasmid vectors contain promoter sequences that drive the transcription of the inserted genes. Different promoter options are available, offering varying levels of gene expression. Regulatory elements, such as enhancers or operators, can be included in plasmid vectors to modulate gene expression and control the timing and magnitude of protein production.
- Size Range: Plasmid vectors can accommodate DNA inserts ranging from a few hundred base pairs to several tens of kilobases. The actual size limit depends on the specific plasmid backbone and the efficiency of DNA insertion. Larger DNA fragments may require alternative vector systems, such as bacterial artificial chromosomes (BACs) or yeast artificial chromosomes (YACs).
- Easy Manipulation: Plasmid vectors are relatively easy to manipulate and modify. They can be easily propagated, isolated, and modified using standard molecular biology techniques. The versatility of plasmid vectors allows for the introduction of specific mutations, deletions, or modifications in the cloned DNA fragments.
- Host Compatibility: Plasmid vectors are primarily used in bacterial host systems, such as Escherichia coli (E. coli). They are well-suited for molecular biology studies and recombinant DNA technology due to the ease of handling and transformation efficiency in bacterial cells. Plasmid vectors can also be used in other host systems, including yeast and mammalian cells, with the necessary modifications.
Plasmid vectors provide a versatile platform for cloning and manipulating DNA fragments. Their ease of use, stability, and compatibility with various host systems make them indispensable tools in molecular biology research, gene expression studies, and recombinant protein production.
2. Bacterial Artificial Chromosomes (BACs)
BACs are large cloning vectors derived from bacterial chromosomes. They have a high capacity to clone large DNA fragments, typically ranging from 100 to 300 kilobases (kb). BACs are often used to clone and maintain large genomic DNA fragments for functional studies, genome mapping, and sequencing projects.
Here are some key features of BACs:
- Cloning Capacity: BACs have a large cloning capacity, allowing for the insertion and maintenance of large DNA fragments. BACs can accommodate DNA inserts ranging from 100 to 300 kilobases (kb) or even larger. This makes them suitable for cloning and studying entire genes, gene clusters, genomic regions, or even small genomes.
- Derived from Bacterial Chromosomes: BACs are derived from the DNA of bacterial chromosomes, typically Escherichia coli (E. coli). They contain elements necessary for replication and stability within the bacterial host. BACs are designed to mimic the behavior of natural bacterial chromosomes, allowing for the stable maintenance and propagation of large DNA inserts.
- Stable Maintenance in Bacteria: BACs are designed to replicate and be stably maintained within bacterial cells. They possess an origin of replication (Ori) sequence derived from the bacterial chromosome, which allows the BAC to replicate along with the host genome during bacterial cell division. This ensures the faithful propagation of the BAC and its DNA insert.
- Cloning Insert Stability: BACs offer high stability for the cloned DNA inserts. The large size of the DNA insert, combined with the elements derived from bacterial chromosomes, helps to prevent rearrangements, deletions, or mutations in the cloned DNA during replication. This stability is important for maintaining the integrity of the cloned DNA for downstream applications.
- Suitable for Genomic Studies: BACs are particularly useful for cloning and studying large genomic fragments. They allow researchers to clone and manipulate entire genes or genomic regions of interest. BAC libraries, which consist of a collection of BAC clones representing the entire genome of an organism, are commonly used for genome mapping, sequencing, and functional genomic studies.
- Compatible with Bacterial Hosts: BACs are compatible with bacterial host systems, primarily E. coli. They can be propagated and manipulated using standard bacterial culture and cloning techniques. E. coli is a well-characterized and easily manipulated organism, making BACs accessible and amenable to various molecular biology techniques.
- Manipulation and Modification: BACs can be easily manipulated and modified using standard molecular biology techniques. Researchers can introduce specific DNA modifications, such as mutations, deletions, or insertions, into the cloned DNA fragments. BACs can also be used for functional studies, such as gene expression analysis or the introduction of reporter genes.
- Genomic Library Construction: BACs are commonly used to construct large-scale genomic libraries. BAC libraries contain a collection of BAC clones, each carrying a different genomic fragment from an organism. These libraries are valuable resources for genome sequencing, mapping, and the identification of genes associated with specific traits or diseases.
3. Yeast Artificial Chromosomes (YACs)
YACs are cloning vectors derived from yeast cells. They can accommodate even larger DNA fragments than BACs, ranging from 100 to 2000 kb. YACs can replicate within yeast cells and are useful for cloning and manipulating large eukaryotic genomes, studying gene regulation, and generating transgenic organisms.
Here are key features of YACs:
- Cloning Capacity: YACs have an exceptionally large cloning capacity, making them suitable for cloning and studying extremely large DNA fragments. YACs can accommodate DNA inserts ranging from 100 kilobases (kb) to 2 megabases (Mb) or even larger. This makes them ideal for cloning and analyzing large genomic regions, including complex gene clusters.
- Derived from Yeast Cells: YACs are derived from the DNA of yeast cells, particularly Saccharomyces cerevisiae. They incorporate elements necessary for replication and stability within the yeast host. YACs are designed to mimic the behavior of natural yeast chromosomes, enabling the stable maintenance and propagation of large DNA inserts.
- Stable Maintenance in Yeast: YACs are designed to replicate and be stably maintained within yeast cells. They contain an origin of replication (Ori) sequence derived from yeast chromosomes, which allows the YAC to replicate alongside the yeast genome during yeast cell division. This ensures the faithful propagation of the YAC and its DNA insert.
- Yeast Compatibility: YACs are compatible with yeast host systems, primarily Saccharomyces cerevisiae. Yeast cells readily take up and propagate YACs, allowing for the manipulation and maintenance of large DNA inserts. Saccharomyces cerevisiae is a well-studied and widely used organism in molecular biology, providing researchers with well-established tools and techniques for working with YACs.
- Mimic Natural Chromosomal Behavior: YACs are designed to behave similarly to natural yeast chromosomes. They possess features that ensure the stability and integrity of the cloned DNA insert during replication and cell division. YACs can undergo normal recombination and segregation, allowing for the study of gene expression, regulation, and genomic interactions in a more natural context.
- Suitable for Genomic Studies: YACs are particularly useful for cloning and studying large genomic fragments. They enable the analysis of entire genes, gene clusters, and large genomic regions, including regulatory elements and complex chromosomal structures. YAC libraries, which consist of a collection of YAC clones representing the entire genome of an organism, are valuable resources for genome mapping, sequencing, and functional genomic studies.
- Manipulation and Modification: YACs can be manipulated and modified using standard molecular biology techniques. Researchers can introduce specific DNA modifications, such as mutations, deletions, or insertions, into the cloned DNA fragments. YACs can also be used for functional studies, such as gene expression analysis or the introduction of reporter genes.
- Stable Maintenance of Large DNA Inserts: YACs provide a stable environment for the maintenance of large DNA inserts. Their large size and natural yeast chromosomal features help prevent rearrangements, deletions, or mutations in the cloned DNA during replication. This stability is crucial for maintaining the integrity and fidelity of the cloned DNA during extended propagation.
4. Viral Vectors
Viral vectors are derived from viruses and are used to introduce foreign DNA into host cells. They have the ability to efficiently deliver DNA into a wide range of cell types. Examples of viral vectors include retroviruses, adenoviruses, lentiviruses, and adeno-associated viruses (AAV). Viral vectors are commonly used in gene therapy, gene expression studies, and genetic engineering experiments.
Here are key features of viral vectors:
- Derived from Viruses: Viral vectors are derived from various types of viruses, including retroviruses, adenoviruses, lentiviruses, adeno-associated viruses (AAV), and others. These viruses have naturally evolved mechanisms for efficient DNA delivery into host cells, which is harnessed for gene transfer in viral vectors.
- Efficient Gene Delivery: Viral vectors are highly efficient in delivering genetic material into host cells. They exploit the natural ability of viruses to enter cells, penetrate the cell membrane, and transfer their genetic cargo into the host cell’s nucleus. This enables the efficient delivery of the cloned DNA or therapeutic genes into target cells.
- Transduction Efficiency: Viral vectors exhibit high transduction efficiency, meaning a large proportion of target cells can take up and express the transferred genes. This efficiency varies depending on the specific viral vector and the target cell type. Viral vectors can transduce both dividing and non-dividing cells, allowing for a wide range of applications.
- Tissue and Cell Type Specificity: Different viral vectors have varying tropism, meaning they have preferences for specific cell types or tissues. This tropism can be natural, based on the properties of the original virus, or engineered to target specific cell types. This feature allows for targeted gene delivery to specific tissues or cell populations.
- Long-Term Gene Expression: Some viral vectors, such as retroviruses and lentiviruses, integrate their genetic material into the host cell’s genome. This integration enables long-term and stable expression of the transferred genes, as they become a part of the host cell’s genetic material. Other viral vectors, like AAV, can persist episomally in the host cell, providing sustained gene expression.
- Different Cargo Capacities: Viral vectors have varying cargo capacities, depending on the size and packaging limitations of the specific virus. Retroviruses and lentiviruses can accommodate relatively larger DNA fragments, while AAV has a more limited capacity. The cargo capacity of viral vectors influences the types of genes or DNA sequences that can be transferred.
- Safety Considerations: Viral vectors undergo extensive safety testing to ensure their use in research and clinical applications. Safety measures include removing or disabling viral genes responsible for replication and pathogenicity, ensuring the inability of the vector to cause disease. Stringent quality control is implemented to minimize the risk of unintended effects or adverse reactions.
- Gene Therapy Applications: Viral vectors are extensively used in gene therapy, which aims to treat genetic disorders by introducing therapeutic genes into affected cells. They have shown promise in correcting genetic defects, delivering missing proteins, or modulating gene expression in target cells. Clinical trials utilizing viral vectors have been conducted for various diseases.
- Gene Expression Studies: Viral vectors are valuable tools in gene expression studies and functional genomics. They enable the overexpression or knockdown of specific genes in target cells, allowing researchers to investigate gene function, signaling pathways, and cellular processes.
- Genetic Engineering: Viral vectors are employed in genetic engineering applications, facilitating the introduction of recombinant DNA constructs into host cells. They enable the creation of genetically modified organisms, the production of recombinant proteins, and the manipulation of gene expression for research or industrial purposes.
5. Phage Vectors
Phage vectors are derived from bacteriophages, which are viruses that infect bacteria. These vectors can carry large DNA fragments and are particularly useful for cloning and manipulating genomic DNA from bacterial species. Phage vectors are advantageous in that they can package and transfer large DNA fragments efficiently into bacterial cells.
Here are key features of phage vectors:
- Bacteriophage Host Specificity: Phage vectors have a specific host range and can infect only certain bacterial species or strains. Different phage vectors are available for different bacterial hosts, such as Escherichia coli (E. coli), Bacillus subtilis, or Pseudomonas aeruginosa. The choice of phage vector depends on the target bacterial host for cloning experiments.
- High Efficiency of Transduction: Phage vectors exhibit high efficiency in transducing bacterial cells with foreign DNA. They have evolved to efficiently infect and transfer their genetic material into host bacteria, making them powerful tools for delivering DNA fragments into bacterial cells.
- Lytic or Lysogenic Life Cycle: Phage vectors can have either a lytic or lysogenic life cycle. Lytic phages immediately infect and replicate within the host bacterium, causing the lysis and destruction of the host cell. Lysogenic phages can integrate their DNA into the bacterial chromosome, establishing a stable relationship where the phage DNA is maintained along with the bacterial genome.
- Cloning Capacity: Phage vectors have a limited cloning capacity compared to other vector types, such as plasmid or BAC vectors. They can accommodate smaller DNA inserts, typically ranging from a few hundred base pairs to a few kilobases. However, the small size of phage vectors can be advantageous in certain applications, such as library construction or high-throughput screening.
- Phage Replication Machinery: Phage vectors carry all the necessary genetic elements for their replication, packaging, and infection into host bacteria. These elements include origin of replication (Ori), packaging signal sequences, and genes encoding proteins required for phage replication. They exploit the host cell’s machinery to produce progeny phages and propagate the inserted DNA fragments.
- Selection and Screening: Phage vectors often incorporate selectable markers, such as antibiotic resistance genes, to allow for the selection of transformed bacterial cells carrying the vector. Additionally, phage vectors can be engineered to include reporter genes or other markers to facilitate the screening of recombinant clones.
- Display Technologies: Phage vectors are commonly used in display technologies, such as phage display or bacterial display systems. These techniques exploit the ability of phages to display peptides or proteins on their surface. By inserting a gene of interest into the phage genome, researchers can express and study a vast library of potential protein interactions or antibody variants.
- Phage Therapy and Antibacterial Applications: Phage vectors have potential applications in phage therapy, where bacteriophages are used to treat bacterial infections. Phage vectors can be engineered to carry genes encoding antibacterial peptides or proteins, allowing for the targeted delivery of antibacterial agents to bacterial pathogens.
- Genetic Manipulation: Phage vectors can be manipulated using molecular biology techniques to introduce specific DNA modifications, insert foreign genes, or delete or mutate existing genes. This enables the study of gene function, regulation, and protein expression in the context of bacterial host systems.
- Ease of Amplification: Phage vectors can be easily amplified in bacterial hosts, allowing for large-scale production of recombinant phages. This is advantageous for applications such as phage display or the production of phage libraries.
6. Cosmid Vectors
Cosmids are hybrid vectors that combine features of plasmids and bacteriophages. They can replicate like plasmids in bacterial cells and package DNA like bacteriophages. Cosmids are useful for cloning DNA fragments of moderate size (up to 45 kb) and are often employed in genomic library construction and DNA sequencing projects.
Here are key features of cosmid vectors:
- Cloning Capacity: Cosmid vectors have a relatively large cloning capacity compared to plasmid vectors. They can accommodate DNA inserts ranging from 35 to 45 kilobases (kb) in size. This makes them suitable for cloning larger DNA fragments, such as genomic DNA or large gene sequences.
- Cos Sites: Cosmid vectors contain the cos site sequences derived from bacteriophage lambda (λ). The cos sites are recognized by the lambda packaging machinery, allowing the cosmid DNA to be packaged into lambda phage particles. This feature enables the efficient packaging and propagation of the cosmid DNA in bacterial cells.
- Origin of Replication (Ori): Cosmid vectors contain a plasmid origin of replication (Ori) sequence. This allows the cosmid DNA to replicate autonomously in bacterial cells, similar to plasmid vectors. The presence of the Ori ensures the stable maintenance and propagation of the cosmid vector and the cloned DNA insert.
- Selectable Markers: Cosmid vectors often include selectable markers, such as antibiotic resistance genes, to enable the selection of bacterial cells carrying the vector. This facilitates the identification and isolation of transformed cells containing the desired cosmid construct.
- Copy Number: Cosmid vectors generally exhibit a moderate to high copy number in bacterial cells. The copy number refers to the number of copies of the cosmid vector present per bacterial cell. Higher copy numbers increase the yield of the cosmid DNA and the cloned DNA insert, which is beneficial for downstream applications.
- Shuttle Vectors: Cosmid vectors can function as shuttle vectors, meaning they can be propagated in both bacterial and eukaryotic cells. This feature allows for the cloning and manipulation of DNA fragments in bacterial hosts and subsequent transfer into eukaryotic systems, such as yeast or mammalian cells.
- DNA Sequencing: Cosmid vectors can be used for DNA sequencing applications. The cloned DNA insert in the cosmid vector can be sequenced using standard DNA sequencing techniques. Cosmid vectors are particularly useful for sequencing larger DNA fragments or genomic regions.
- Genomic Library Construction: Cosmid vectors are commonly used in the construction of genomic libraries. Genomic libraries are collections of DNA fragments representing an organism’s entire genome. The large cloning capacity of cosmid vectors allows for the construction of comprehensive genomic libraries, which are valuable resources for genomic research.
- DNA Manipulation: Cosmid vectors can be manipulated using molecular biology techniques to introduce specific DNA modifications, insert foreign genes, or delete or mutate existing genes. This enables the study of gene function, regulation, and protein expression using cosmid-based constructs.
- Cosmid-λ Hybrid System: Cosmid vectors can be combined with lambda (λ) vectors to create a cosmid-λ hybrid system. This system allows for the efficient packaging and propagation of larger DNA fragments than what can be accommodated by cosmid vectors alone. The hybrid system combines the cloning capacity of cosmid vectors with the packaging capacity of lambda phage particles.
7. Expression Vectors
Expression vectors are specifically designed for the expression of foreign genes in host cells. They contain regulatory elements such as promoters, enhancers, and terminators that enable efficient transcription and translation of the inserted gene. Expression vectors are widely used in recombinant protein production, functional studies of genes, and protein engineering.
- Promoter: Expression vectors include a promoter region, which is responsible for initiating the transcription of the gene of interest. Promoters can be constitutive, driving gene expression continuously, or inducible, allowing for control of gene expression based on specific conditions or stimuli.
- Enhancer Elements: Expression vectors may contain enhancer elements that enhance the activity of the promoter, resulting in increased gene expression levels. Enhancers are regulatory DNA sequences that interact with transcription factors to facilitate efficient transcriptional activation.
- Multiple Cloning Site (MCS): Expression vectors have a multiple cloning site or polylinker region, which is a short DNA sequence that contains multiple unique restriction enzyme recognition sites. This allows for the convenient insertion of the gene of interest into the vector at the desired location.
- Transcription Termination Sequence: Expression vectors contain a transcription termination sequence that signals the end of transcription and ensures proper processing of the mRNA transcript. This sequence helps to generate a stable mRNA molecule for subsequent translation.
- Ribosome Binding Site (RBS): Expression vectors designed for bacterial expression often contain a ribosome binding site (RBS) upstream of the start codon. The RBS facilitates efficient translation initiation by positioning the ribosome at the appropriate site on the mRNA molecule.
- Translation Initiator: Expression vectors include a translation initiator, typically a start codon (usually AUG), which signals the beginning of translation. This codon is located immediately upstream of the coding sequence of the gene of interest.
- Selectable Markers: Expression vectors commonly incorporate selectable markers, such as antibiotic resistance genes, which enable the selection and identification of host cells that have successfully taken up and expressed the vector. Selectable markers facilitate the isolation of cells with the desired expression construct.
- Fusion Tags: Expression vectors may contain fusion tags or reporter genes that can be attached to the gene of interest. These tags allow for easy detection, purification, or localization of the expressed protein. Examples of fusion tags include green fluorescent protein (GFP), His-tag, GST-tag, or FLAG-tag.
- Secretion Signals: Some expression vectors are designed for the secretion of the expressed protein into the extracellular space or periplasmic space of the host organism. These vectors contain secretion signals or leader sequences that direct the protein to the appropriate cellular compartment for secretion.
- Host Compatibility: Expression vectors are available for a wide range of host organisms, including bacteria (e.g., E. coli), yeast (e.g., Saccharomyces cerevisiae), insect cells (e.g., baculovirus-based systems), mammalian cells, and others. The choice of expression vector depends on the specific host organism and its compatibility with the vector system.
Other Types of Vectors;
- Transcription Vectors: Transcription vectors are specialized vectors designed for studying transcriptional regulation and analyzing promoter activity. They typically contain reporter genes, such as luciferase or β-galactosidase, that allow for the measurement of transcriptional activity. Transcription vectors are valuable tools for investigating gene expression patterns, identifying regulatory elements, and studying transcriptional control mechanisms.
- Human Artificial Chromosomes (HACs): Human artificial chromosomes are vectors that are engineered to mimic the structure and behavior of natural human chromosomes. HACs can carry large DNA fragments, ranging from hundreds of kilobases to megabases in size, making them suitable for studying large genomic regions and gene therapy applications. HACs have the potential to provide stable, long-term expression of genes in human cells without the risk of integration into the host genome.
- Bacteriophage Vectors: Bacteriophages are viruses that infect bacteria, and their genomes can be engineered to serve as cloning vectors. Phage vectors are commonly used for cloning and manipulating DNA fragments in bacterial hosts. They offer advantages such as high cloning efficiency, rapid propagation, and easy purification. Examples of bacteriophage vectors include lambda phage vectors (λ vectors) and M13 phage vectors.
- Adenoviral Vectors: Adenoviral vectors are derived from adenoviruses, which are non-integrating DNA viruses that can efficiently infect a wide range of dividing and non-dividing cells. Adenoviral vectors are commonly used for gene transfer and gene therapy applications. They can accommodate relatively large DNA inserts and provide transient gene expression in mammalian cells without integrating into the host genome.
- Retroviral Vectors: Retroviral vectors are derived from retroviruses, which are RNA viruses that can integrate their genetic material into the host cell genome. Retroviral vectors are widely used for stable gene transfer and long-term gene expression in both dividing and non-dividing cells. They are valuable tools in gene therapy, stem cell research, and genetic engineering experiments.
- Lentiviral Vectors: Lentiviral vectors are a type of retroviral vectors that are derived from lentiviruses, such as the human immunodeficiency virus (HIV). Lentiviral vectors can efficiently deliver DNA into both dividing and non-dividing cells, including stem cells. They are commonly used for stable gene transfer, gene knockdown, and other genetic manipulation experiments.
| Cloning Vector | Key Features |
|---|---|
| Plasmid Vectors | Circular DNA molecules capable of autonomous replication |
| Multiple cloning sites for easy insertion of DNA fragments | |
| Selection markers for identifying transformed cells | |
| Varying copy numbers (high or low) depending on the origin | |
| Bacterial Artificial Chromosomes (BACs) | Derived from bacterial chromosomes |
| Large cloning capacity (100-300 kb) for genomic fragments | |
| Stable maintenance and propagation in bacteria | |
| Suitable for genome mapping, sequencing, and functional studies | |
| Yeast Artificial Chromosomes (YACs) | Derived from yeast cells |
| Very large cloning capacity (100-2000 kb) for eukaryotic genomes | |
| Capable of replicating within yeast cells | |
| Ideal for cloning large genomic fragments and studying gene regulation | |
| Viral Vectors | Derived from viruses for efficient DNA delivery into cells |
| Retroviruses, adenoviruses, lentiviruses, AAV, etc. | |
| High transduction efficiency in a wide range of cell types | |
| Used for gene therapy, gene expression, and genetic engineering | |
| Phage Vectors | Derived from bacteriophages (viruses that infect bacteria) |
| Efficient packaging and transfer of large DNA fragments | |
| Commonly used for cloning and manipulating bacterial genomic DNA | |
| Cosmid Vectors | Hybrid vectors combining features of plasmids and phages |
| Replicate like plasmids in bacterial cells | |
| Package DNA like bacteriophages | |
| Suitable for cloning DNA fragments up to 45 kb | |
| Expression Vectors | Designed for efficient expression of genes in host cells |
| Include specific promoters, enhancers, and terminators | |
| Enable high levels of transcription and translation | |
| Widely used in recombinant protein production and functional studies | |
| Transcription Vectors | Specialized vectors for studying transcriptional regulation |
| Contain reporter genes for measuring transcriptional activity | |
| Used to investigate gene expression patterns and regulatory elements | |
| Human Artificial Chromosomes (HACs) | Engineered to mimic natural human chromosomes |
| Can carry large DNA fragments (100s of kb to Mb) for gene therapy | |
| Provide stable, long-term expression without integration | |
| Used for genomic studies, gene delivery, and therapeutic applications | |
| Bacteriophage Vectors | Derived from bacteriophages for cloning and manipulating DNA |
| Efficiently package and transfer DNA into bacterial cells | |
| Used for cloning, library construction, and genetic engineering |
Differences Between Different Types of Cloning Vectors
| Vector Type | Cloning Capacity | Host Range | Copy Number | Promoters | Selectable Markers | Expression Features | Applications |
|---|---|---|---|---|---|---|---|
| Plasmid Vectors | Small to Medium | Broad | High | Varied | Antibiotic Resistance | Limited expression | Gene cloning, gene expression, protein production |
| Bacterial Artificial Chromosomes (BACs) | Large | Bacteria | Low to Moderate | Varied | Antibiotic Resistance | Limited expression | Large DNA cloning, genomic libraries |
| Yeast Artificial Chromosomes (YACs) | Very Large | Yeast | Low to Moderate | Varied | Auxotrophic markers | Moderate expression | Large DNA cloning, genomic libraries |
| Viral Vectors | Variable | Varies by viral type | Variable | Viral promoters | Viral or Antibiotic | High expression | Gene delivery, gene therapy, vaccine development |
| Phage Vectors | Small to Medium | Bacteria | Moderate to High | Bacterial promoters | Antibiotic Resistance | Limited expression | Bacterial cloning, phage display, library construction |
| Cosmid Vectors | Medium | Bacteria | Moderate | Varied | Antibiotic Resistance | Limited expression | Large DNA cloning, genomic libraries |
| Expression Vectors | Varied | Varied | Varied | Strong promoters | Antibiotic Resistance | High expression | Gene expression, protein production, functional analysis |
Advanatages of Cloning Vectors
- DNA Amplification: Cloning vectors allow for the amplification of DNA fragments of interest through replication in host cells. This enables the generation of large quantities of DNA for further analysis, manipulation, and experimentation.
- Versatility: Cloning vectors are available in a variety of types and sizes, allowing researchers to choose the most suitable vector for their specific needs. Different vectors offer various features and functionalities, such as the capacity to carry large DNA inserts, specific promoter sequences, or reporter genes, providing flexibility in experimental design.
- Gene Manipulation: Cloning vectors facilitate the manipulation of genes, including the introduction of mutations, deletions, or modifications. This enables researchers to study gene function, protein structure, and regulatory elements. Cloning vectors also allow for the construction of fusion proteins and the addition of molecular tags to facilitate protein detection and analysis.
- Expression Systems: Cloning vectors are designed to drive the expression of genes in host cells, allowing for the production of recombinant proteins. They provide the necessary elements, such as promoters and regulatory sequences, for efficient transcription and translation of the inserted genes. This enables the study of protein function, purification, and therapeutic protein production.
- Genetic Libraries: Cloning vectors are used to construct DNA libraries, which contain large collections of DNA fragments or genes. Libraries facilitate genome mapping, gene discovery, and functional genomics studies. Cloning vectors enable the propagation and maintenance of DNA fragments in a format suitable for high-throughput screening and analysis.
Limitations of Cloning Vectors
- Insert Size Limit: Cloning vectors have limitations on the size of DNA fragments they can accommodate. Some vectors have size restrictions, which may hinder the cloning of large genes or genomic regions. For cloning large DNA fragments, specialized vectors such as bacterial artificial chromosomes (BACs) or yeast artificial chromosomes (YACs) may be required.
- Transformation Efficiency: The efficiency of cloning vector uptake and transformation into host cells can vary. Some vectors may have lower transformation efficiencies, resulting in a smaller number of successful transformations. This can affect the yield of recombinant clones and the efficiency of downstream applications.
- Vector-Host Compatibility: Cloning vectors are designed for specific host organisms, such as bacteria, yeast, or mammalian cells. Compatibility issues may arise when attempting to use a cloning vector in a non-optimal host system, leading to lower efficiency or lack of replication. It is essential to choose the appropriate vector for the specific host organism of interest.
- Stability and Copy Number: The stability and copy number of cloning vectors can vary. Some vectors may exhibit instability or low copy number in host cells, which can affect the maintenance and propagation of the inserted DNA fragment. Researchers need to consider vector stability when selecting a suitable vector for their experiments.
- Sequencing Artifacts: Cloning vectors used in DNA sequencing may introduce artifacts or biases during the sequencing process. These artifacts can result from the vector sequences or the cloning process itself and may affect the accuracy or interpretation of sequencing results. It is important to consider and account for any potential artifacts associated with the specific cloning vector used.
Uses of Cloning Vectors
- Gene Cloning: Cloning vectors are extensively used for the cloning of genes or DNA fragments of interest. By inserting the DNA fragment into the vector, researchers can replicate and propagate the DNA in host cells, allowing for further analysis, manipulation, and study.
- Recombinant Protein Expression: Cloning vectors play a crucial role in the expression of recombinant proteins. They provide the necessary elements, such as promoters, regulatory sequences, and selectable markers, to enable the efficient transcription and translation of the gene of interest in host cells. Recombinant protein expression is used in various applications, including protein purification, structural studies, functional characterization, and therapeutic protein production.
- Molecular Tagging and Fusion Proteins: Cloning vectors allow for the fusion of genes or DNA fragments with specific tags or reporter genes. This enables the visualization, purification, or functional analysis of the tagged protein. Common tags include green fluorescent protein (GFP), His-tag, FLAG-tag, and Myc-tag, which facilitate protein detection, localization, and interaction studies.
- Functional Studies and Mutagenesis: Cloning vectors are used to introduce mutations or modifications into genes of interest, enabling the study of gene function and structure. Techniques such as site-directed mutagenesis, gene deletion, or gene knockdown can be performed using cloning vectors to investigate the effects of specific genetic changes on protein function or cellular processes.
- DNA Library Construction: Cloning vectors are essential for the construction of DNA libraries, which contain large collections of DNA fragments or genes. Libraries can be used for various purposes, such as genome mapping, gene discovery, functional genomics, and drug target identification. Cloning vectors allow the insertion and maintenance of DNA fragments in a format suitable for high-throughput screening and analysis.
- Gene Therapy: Cloning vectors, particularly viral vectors, are widely used in gene therapy applications. They serve as vehicles to deliver therapeutic genes into target cells or tissues, aiming to correct genetic disorders, treat diseases, or provide therapeutic benefits. Viral vectors, such as retroviruses and adenoviruses, efficiently deliver genes into cells for long-term expression and therapeutic effect.
- DNA Sequencing: Cloning vectors are used in DNA sequencing workflows. They can carry and propagate DNA fragments of interest, which can then be isolated and sequenced. Cloning vectors provide a means to amplify and maintain the DNA fragments during the sequencing process, enabling the generation of sufficient amounts of DNA for analysis.
Topo Cloning Vectors – What is it?
Topo cloning vectors, also known as topoisomerase-based cloning vectors, are a specific type of cloning vector used in molecular biology research. They utilize the activity of topoisomerase enzymes to facilitate the cloning of DNA fragments into the vector.
Topoisomerases are enzymes that can modify the topology of DNA by breaking and rejoining the DNA strands, allowing for the relaxation of supercoiled DNA. Topo cloning vectors are designed to take advantage of the activity of topoisomerases to simplify the cloning process. The most commonly used topo cloning vector is the pCR®2.1-TOPO® vector, which is commercially available.
The key features and steps involved in topo cloning vectors are as follows:
- Topoisomerase Recognition Site: Topo cloning vectors contain a specific recognition site for the topoisomerase enzyme. The recognition site consists of a short sequence, typically 3-5 base pairs, which is specific to the particular topoisomerase used.
- Topoisomerase-Mediated Cloning: The DNA fragment of interest, typically generated by PCR amplification, is mixed with the linearized topo cloning vector. The linearized vector contains the topoisomerase recognition site at each end.
- Topoisomerase Reaction: The topoisomerase enzyme recognizes and binds to the recognition site in the vector and forms a covalent bond with the DNA fragment. This results in the formation of a covalently closed circular DNA molecule that includes the vector and the inserted DNA fragment.
- Transformation: The covalently closed circular DNA molecule is then introduced into a host organism, such as bacteria, through a process called transformation. The transformed bacteria take up and propagate the topo cloning vector.
- Selection and Screening: The topo cloning vector typically includes selectable markers, such as antibiotic resistance genes, which allow for the selection and identification of bacteria that have successfully taken up the vector. Screening methods, such as restriction enzyme digestion or sequencing, are used to confirm the presence and integrity of the cloned DNA fragment.
Topo cloning vectors offer several advantages in the cloning process. They are simple to use and do not require the use of restriction enzymes for DNA fragment insertion, reducing the risk of unintended side reactions. Additionally, the use of topoisomerases allows for efficient cloning of both blunt-ended and cohesive-ended DNA fragments, providing flexibility in the choice of DNA sources.
In summary, topo cloning vectors employ topoisomerase enzymes to facilitate the cloning of DNA fragments into the vector. They provide a convenient and efficient method for the insertion of DNA fragments into circular vectors, simplifying the cloning process in molecular biology research.
Transposons are frequently used as cloning vectors.
Transposons, also known as transposable elements, are segments of DNA that have the ability to move around within a genome. They can be found in the genomes of various organisms, including bacteria, plants, and animals. While transposons were initially considered “junk DNA” with no clear function, they have been found to play important roles in genome evolution and gene regulation.
In the context of cloning, transposons can be utilized as cloning vectors. Cloning vectors are DNA molecules used to carry foreign DNA fragments into host organisms, where they can be replicated and expressed. Transposons can serve as effective cloning vectors due to their ability to insert themselves into the host genome.
Here’s a simplified overview of how transposons can be used as cloning vectors:
- Selection of a transposon: A transposon with known characteristics and a well-understood mechanism of transposition is chosen as the cloning vector. Examples of commonly used transposons include Tn5 and Tn10 in bacteria.
- Insertion of desired DNA: The transposon vector is modified by inserting the desired DNA fragment, which typically contains a gene of interest, into the transposon DNA sequence. This is often achieved through DNA recombinant techniques.
- Transformation or transfection: The modified transposon vector is introduced into the host organism (e.g., bacteria, yeast, or cultured cells) using transformation (in the case of bacteria) or transfection (in the case of eukaryotic cells).
- Transposition and integration: Once inside the host organism, the transposon vector undergoes transposition, a process in which the transposon DNA segment moves from the vector to a new location within the host genome. This results in the integration of the desired DNA fragment into the host genome.
- Selection of transformed cells: To identify host cells that have successfully taken up the transposon vector and incorporated the desired DNA fragment, selection markers such as antibiotic resistance genes or fluorescent proteins are often included in the vector. Only the transformed cells that contain the transposon vector will survive under specific selection conditions.
- Verification and propagation: The transformed cells are further analyzed to confirm the presence and proper integration of the desired DNA fragment. Once verified, the transformed cells can be propagated, allowing the cloned DNA to be replicated and expressed.
Using transposons as cloning vectors offers several advantages. They can carry relatively large DNA fragments, including entire genes or gene clusters. Additionally, transposons can integrate the DNA fragment into a specific site or randomly throughout the host genome, depending on the transposon used and the experimental design. This versatility allows for various applications, including gene expression studies, mutagenesis, and functional genomics research.
It’s worth noting that while transposons can be valuable tools for cloning, other vector systems, such as plasmids and viral vectors, are also commonly used depending on the specific requirements of the cloning experiment.
FAQ
What is a cloning vector, and what is its purpose?
A cloning vector is a DNA molecule that is used to carry and replicate foreign DNA fragments in a host organism. Its purpose is to enable the amplification and manipulation of specific DNA sequences.
How are cloning vectors different from other types of vectors?
Cloning vectors are specifically designed for the cloning and propagation of DNA fragments, while other types of vectors, such as expression vectors or viral vectors, have additional features tailored for specific purposes like gene expression or gene delivery.
What are the key features to consider when selecting a cloning vector?
Important features to consider include cloning capacity (size of DNA fragments it can accommodate), selectable markers, copy number, compatibility with host organisms, and expression features if gene expression is desired.
What are the common selectable markers used in cloning vectors?
Selectable markers are genes that confer resistance to antibiotics or provide other means of selection, such as auxotrophic markers in yeast. Examples include ampicillin, kanamycin, or neomycin resistance genes.
Can I use the same cloning vector for different host organisms?
Some cloning vectors are versatile and can be used in various host organisms, while others are specific to certain hosts. It is important to choose a vector compatible with the desired host organism to ensure efficient replication and expression.
Can cloning vectors accommodate large DNA fragments?
The cloning capacity of vectors varies. Plasmid vectors can typically accommodate small to medium-sized DNA fragments, while BACs (Bacterial Artificial Chromosomes) or YACs (Yeast Artificial Chromosomes) are suitable for larger DNA fragments.
Are there different types of promoters available in cloning vectors?
Yes, cloning vectors can offer a range of promoters, including constitutive promoters for continuous expression or inducible promoters that allow controlled gene expression under specific conditions.
How can I ensure proper replication and stability of the cloning vector?
Cloning vectors contain an origin of replication (Ori) sequence that ensures their autonomous replication within the host organism. Proper selection of the vector and appropriate culture conditions help maintain stability.
Can I modify the cloning vector to suit my experimental needs?
Yes, cloning vectors can be modified through genetic engineering techniques. The introduction or removal of specific DNA sequences, such as promoters, terminators, or fusion tags, allows customization of the vector for specific applications.
What are the limitations of cloning vectors?
Cloning vectors have limitations in terms of cloning capacity, expression levels, or compatibility with certain host organisms. Additionally, the presence of the vector itself may affect the biology of the host organism in some cases. These factors need to be considered when selecting a suitable cloning vector.
References
- Del S.G et al. (1998). Replication and control of circular bacterial plasmids. Microbiology and molecular biology reviews. 62(2): 434-464
- Castagnoli. L et al. (1989). Genetic and structural analysis of the ColE1 Rop (Rom) protein. EMBO J. 8(2): 621-629
- Feinbaum.R (2001). Introduction to plasmid biology. Current protocols in molecular biology. 1-5
- Vocke. C and Bastia. D. (1983). Primary structure of the essential replicon of the plasmid pSC101. PNAS. 80.21: 6557-6561
- https://www.sciencedirect.com/topics/neuroscience/cloning-vector
- https://www.khanacademy.org/science/in-in-class-12-biology-india/xc09ed98f7a9e671b:in-in-biotechnology-principles-and-processes/xc09ed98f7a9e671b:restriction-digests-cloning-and-transformation/a/features-of-cloning-vectors
- https://goldbio.com/articles/article/Cloning-Vectors-Essential-Features
- https://www.snapgene.com/plasmids/basic_cloning_vectors