What is BLAST?
BLAST, which stands for Basic Local Alignment Search Tool, is a widely used bioinformatics program and algorithm. It is designed to compare and analyze biological sequences such as DNA, RNA, and protein sequences. BLAST helps in identifying regions of similarity between different sequences, which can provide insights into their functional and evolutionary relationships.
BLAST works by searching a query sequence against a database of known sequences. It employs a heuristic algorithm to quickly identify local alignment regions that exhibit significant similarity between the query and database sequences. BLAST calculates a statistical score, known as the E-value, which indicates the probability of finding a similar sequence by chance.
The results of a BLAST search provide information about the matches found in the database, including the degree of similarity, alignment scores, and the locations of matches. BLAST has numerous applications in various fields of biological research, such as genome annotation, protein structure prediction, gene identification, and phylogenetic analysis.
There are different variants of BLAST available, each optimized for specific types of sequences and search requirements. Some of the commonly used variants include BLASTn for nucleotide-nucleotide comparisons, BLASTp for protein-protein comparisons, BLASTx for translating nucleotide sequences to protein sequences, and tBLASTn/tBLASTx for comparing translated nucleotide sequences against protein databases.
Types of BLAST
There are a variety of BLAST algorithms, each of which is designed for a particular form of sequence comparison. Here are the five most common varieties of BLAST:
- BLASTn: BLASTn is utilized to compare nucleotide sequences to a nucleotide database. Primarily, it is used to identify similarities and locate homologous regions in DNA sequences.
- BLASTp: BLASTp is used to compare protein sequences against a database of protein sequences. It facilitates the identification of similar protein sequences, which can shed light on protein function, structure, and evolution.
- BLASTx: BLASTx is used to compare a nucleotide query sequence to a database of proteins. It translates the query DNA sequence in all six reading frames and compares the resultant amino acid sequences to the database of proteins. BLASTx is particularly effective when searching DNA sequences for protein-coding genes.
- tBLASTn: tBLASTn compares a protein query sequence to a nucleotide database. It translates the nucleotide database sequences in each of the six reading frames and then compares the resulting amino acid sequences to the protein query. When searching for potential protein homologs in DNA sequences, tBLASTn is frequently employed.
- tBLASTx: tBLASTx is a combination of translated nucleotide (DNA) queries and searches of translated nucleotide (DNA) databases. It translates both the query and database sequences in all six reading frames, compares the resulting amino acid sequences, and provides information regarding any possible similarities. When searching for similarities between two DNA nucleotide sequences, tBLASTx is frequently utilized.
Special kinds of BLASTs
In addition to the standard BLAST algorithms (BLASTn, BLASTp, BLASTx, tBLASTn, tBLASTx), there are several special kinds of BLASTs that have been developed to address specific needs in sequence analysis. Here are a few examples:
- PSI-BLAST (Position-Specific Iterated BLAST): PSI-BLAST is an iterative version of BLASTp that aims to improve the detection of distantly related protein sequences. It builds a position-specific scoring matrix (PSSM) based on the alignments found in previous iterations, allowing for the identification of more divergent homologs.
- PHI-BLAST (Pattern-Hit Initiated BLAST): PHI-BLAST is used for identifying and aligning protein sequences that contain specific patterns or motifs. It starts with a pattern search against a protein database and then extends the search using a BLAST-like algorithm.
- DELTA-BLAST: DELTA-BLAST is a tool that combines the advantages of PSI-BLAST and HMMER. It performs a search using PSI-BLAST and then uses the identified homologs to construct a position-specific scoring matrix (PSSM), which is further used to perform a search with HMMER against a custom database.
- Reverse-BLAST: Reverse-BLAST is used to find potential nucleotide or protein sources for a given sequence. It takes a query sequence and searches for its potential sources by comparing it against a database of known sequences.
- Short-Read BLAST (SR-BLAST): SR-BLAST is designed to handle the analysis of short DNA sequence reads generated by high-throughput sequencing technologies. It allows for efficient and accurate alignment of short reads against a reference database, enabling tasks such as read mapping and variant calling.
These specialized BLAST variants cater to specific research needs and provide enhanced capabilities for analyzing different types of sequences or addressing specific sequence analysis challenges.
Characteristics of BLAST
BLAST (Basic Local Alignment Search Tool) possesses a number of essential characteristics that contribute to its efficacy and pervasive application in sequence analysis. Here are some distinguishing features of BLAST:
- Speed and Efficiency: BLAST is designed to perform sequence similarity searches quickly and efficiently. It utilizes heuristic algorithms and indexing techniques to expedite the identification of local alignments, making it suitable for searching large sequence databases in a reasonable amount of time.
- Sensitivity and Specificity: In sequence comparisons, BLAST establishes a balance between sensitivity and specificity. It seeks to identify meaningful correlations while minimizing false positives. BLAST provides a measure of the statistical significance of the identified matches by utilizing scoring matrices and statistical measures such as E-values.
- Focus on Local Alignments: BLAST focuses on identifying local rather than global alignments. It identifies shorter regions of substantial similarity, known as high-scoring segment pairs (HSPs), which enables efficient identification of conserved regions even in sequences with divergent characteristics.
- Iterative Method: Some BLAST variants, such as PSI-BLAST, employ an iterative method. They conduct multiple cycles of searching and alignment to refine the query and database sequences progressively. This iterative procedure facilitates the detection of more distant homologs and increases sensitivity.
- Flexibility: BLAST is versatile and can be applied to numerous categories of biological sequences, such as DNA, RNA, and proteins. Different BLAST variants are tailored to specific sequence types and search criteria, allowing for versatility in sequence analysis duties.
- User-Friendly Interface: BLAST tools typically feature user-friendly interfaces that enable researchers to readily input query sequences, select databases, and configure search parameters. This accessibility enables users with differing degrees of bioinformatics knowledge to conduct efficient sequence similarity searches.
- Extensive Database Compatibility: BLAST is compatible with a vast array of sequence databases, including public databases such as GenBank, UniProt, and the NCBI’s non-redundant (nr) database. This compatibility enables researchers to compare their sequences to exhaustive collections of previously identified sequences.
- Community Support and Updates: BLAST has a sizable user community, which has aided in its ongoing development and updates. Regular updates and issue fixes ensure that BLAST remains a trustworthy and current sequence analysis tool.
How BLAST Works
- The BLAST algorithm is a heuristic program, which means it uses intelligent shortcuts to perform the search more quickly.
- BLAST performs “local” alignments. Functional domains are frequently repeated within the same protein as well as across proteins from different species in the vast majority of proteins.
- The BLAST algorithm is optimized to identify these domains or shorter sequence-similar segments. Local alignment also allows an mRNA to be aligned with a fragment of genomic DNA, which is frequently necessary for genome assembly and analysis.
- If BLAST initially attempted to align two sequences along their entire lengths (known as a global alignment), fewer similarities would be detected, particularly in terms of domains and motifs.
- When a query is submitted through one of the BLAST Web pages, the sequence, along with any other input information such as the database to be searched, word size, expected value, etc., is supplied to the algorithm on the BLAST server.
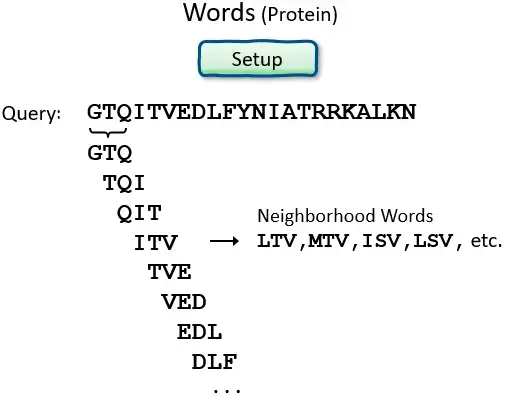
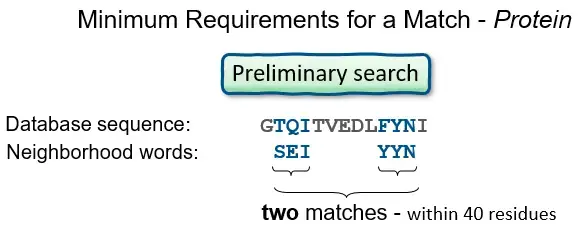
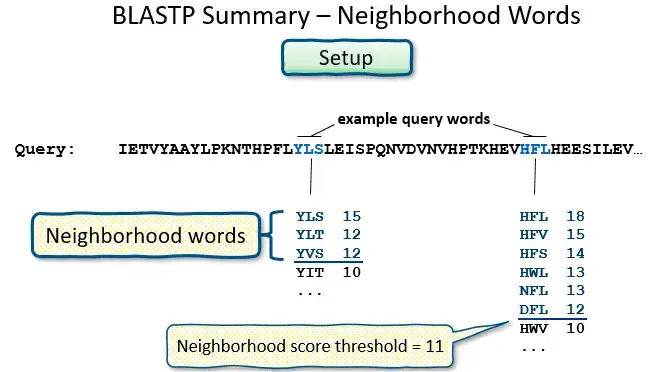
- BLAST operates by first creating a look-up table of all the “words” (brief subsequences, which for proteins have a default length of three letters) and “neighboring words,” i.e., words in the query sequence that are similar to the query words.
- The sequence database is then searched for these “hot spots” When a match is found, it is utilized to generate gap-free and gapped extensions of the “word.” Directly searching GenBank flatfiles (or any subset of GenBank flatfiles) is not supported by BLAST.
- Sequences are instead added to BLAST databases. Each entry is divided into two files, one containing only the header information and the other containing only the sequence information.
- These are the data utilized by the algorithm. If BLAST is to be executed in “stand-alone” mode, the data file may contain local, private data, downloaded NCBI BLAST databases, or a combination of both.
- After the algorithm has searched for and maximally extended all possible “words” from the query sequence, it assembles the best alignment for each query–sequence pair and writes this information to a SeqAlign data structure. The SeqAlign structure does not contain sequence information; instead, it references the sequences in the BLAST database.
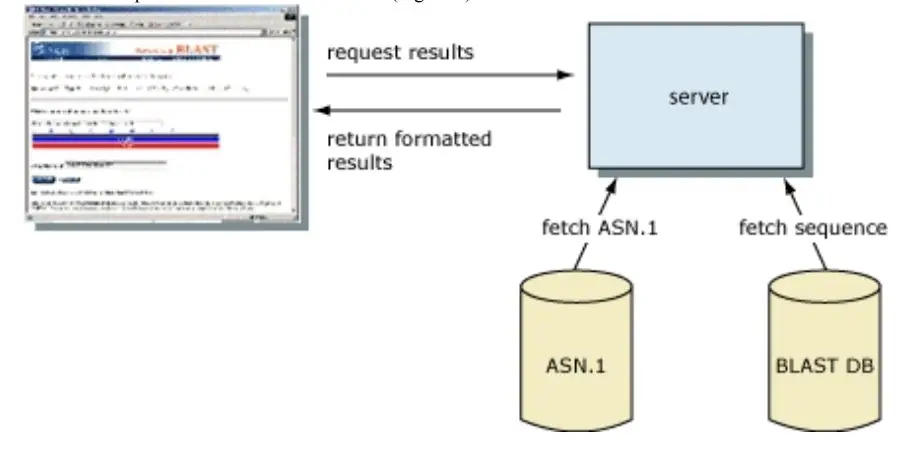
- The BLAST Formatter, which resides on the BLAST server, can utilize the information in the SeqAlign to retrieve and display similar sequences in a variety of ways. Therefore, once a query has been executed, the results can be reformatted without rerunning the search. This is made feasible by the QBLAST system.
BLAST Scores and Statistics
- Once BLAST has identified a similar sequence to the query in the database, it is useful to determine whether the alignment is “good” and whether it depicts a possible biological relationship, or whether the similarity observed is due to chance alone.
- BLAST employs statistical theory to generate a bit score and expect value (E-value) for each alignment pair (query to match) using statistical theory. The bit score indicates the quality of the alignment; the higher the score, the higher the quality of the alignment. In general, this score is computed using a formula that considers the alignment of similar or identical residues, as well as any voids introduced to align the sequences.
- The “substitution matrix,” which assigns a score for aligning any possible pair of residues, is a crucial component of this calculation. The exceptions to this are blastn and MegaBLAST, which perform nucleotide–nucleotide comparisons and therefore do not use protein-specific matrices.
- Bit scores are normalized, allowing bit scores from various alignments to be compared despite the use of different scoring matrices. The E-value indicates the statistical significance of a given pairwise alignment and reflects the database size and scoring system employed.
- Lesser the E-value, greater the significance of the impact. An E-value of 0.05 for a sequence alignment indicates that this similarity has a 5 in 100 (1 in 20) probability of occurring by chance alone.
- Although a statistician may consider this to be significant, it may not represent a biologically meaningful result; an alignment analysis (see below) is required to ascertain “biological” significance.
BLAST Output
1. The Traditional Report
The majority of BLAST users are acquainted with the “traditional” BLAST report. The report is divided into three sections: (1) the database header, which comprises information about the query sequence. On the Internet, there is also a graphical overview; (2) one-line descriptions of each database sequence found to match the query sequence; these provide a quick overview for browsing; and (3) alignments for each database sequence matched (there may be multiple alignments for a database sequence it matches).
The BLAST report header
The first line contains information regarding the program classification (in this case, BLASTP), version (2.2.1), and version release date. The paper describing BLAST is then cited, followed by the request ID (issued by QBLAST), the query sequence definition line, and a summary of the database that was searched. The Taxonomy reports link displays this BLAST result based on Taxonomy database information.

Graphical overview of BLAST results
The numbered red bar at the top of the figure depicts the query sequence. Below the red bar, database results aligned with the query are displayed. The most similar aligned sequences are displayed closest to the query. In this instance, three database matches with high scores align with the majority of the query sequence. The subsequent twelve bars represent lower-scoring matches that align to two regions of the query, between residues 3 and 60 and residues 220 and 500. The crosshatched portions of these bars indicate that the two regions of similarity are located on the same protein, but that the region in between does not match. The remaining bars represent alignments with lower scores. When the mouse is hovered over the bars, the sequence’s definition line is displayed in the window located above the graphic.

One-line descriptions in the BLAST report
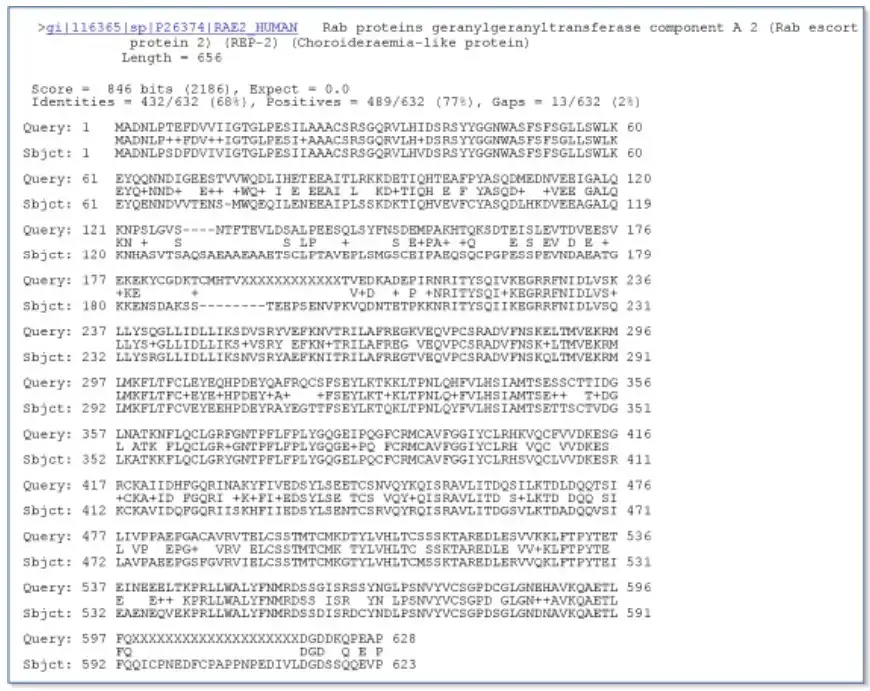
(a) the gi number, database designation, Accession number, and locus name for the matched sequence, separated by vertical bars (Appendix 1); (b) a concise textual description of the sequence, the definition. This typically includes information about the organism from which the sequence was derived, the type of sequence (e.g., mRNA or DNA), and some function or phenotype information. To maintain a compact display, the definition line is frequently truncated in one-line descriptions; (c) the alignment score in bits. The results with the highest scores appear at the head of the list; (d) the E-value, which provides an estimate of statistical significance. For the first finding on the list, the gi number is 116365, the database designation is sp (for SWISS-PROT), the Accession number is P26374, the locus name is RAE2_HUMAN, the score is 1216, and the E-value is 0.0. Note that the first seventeen hits have extremely low E-values (significantly less than 1) and are either RAB proteins or GDP dissociation inhibitors. The other database matches have E-values of 0.5 or higher, indicating that these sequences may have matched by coincidence alone.

A pairwise sequence alignment from a BLAST report
The sequence identifier, the complete definition line, and the length of the matched sequence in amino acids precede the alignment. The bit score (the original score is enclosed in parentheses) and the E-value follow. The line that follows contains information regarding the number of identical residues in this alignment (Identities), the number of conservative substitutions (Positives), and, if applicable, the number of alignment gaps. The actual alignment is then displayed, with the query on top and the database match designated Sbjct below. The numbers on the left and right indicate the position within the sequence of amino acids. One or more dashes (–) denote insertions or deletions within a sequence. Xs are substituted for amino acid residues in the query sequence that have been masked due to their minimal complexity (see, for example, the fourth and final blocks). The line between the two sequences indicates the sequences’ similarities. If the query and the subject contain the same amino acid at a particular position, the residue itself is displayed. Conservative substitutions, as determined by the substitution matrix, are denoted by a plus sign.
- The conventional report is intended for human consumption as opposed to programmatic parsing. One-line descriptions, for instance, are helpful for gaining a fast overview of search results, but due to limited space, they are rarely exhaustive.
- Also, several bits of information (such as the E-values, scores, and descriptions) are displayed in both the one-line descriptions and alignments for convenience; thus, the person viewing the search output does not need to toggle between sections.
- New features, such as the inclusion of links to Entrez Gene records from sequence hits, may be added to the report, resulting in a change of output format. These are simple to recognize and exploit, but they can upset programs that parse this BLAST output.
- On the advanced BLAST page, the Alignments option can be used to alter the default maximum of 500 sequence matches that are displayed.
- Numerous components of the BLAST results are hyperlinked to the same information in multiple locations on the page, to additional information including assistance documentation, and to the Entrez sequence records of matched sequences. These records provide additional information regarding the sequence, including links to relevant PubMed abstracts.
2. The Hit Table
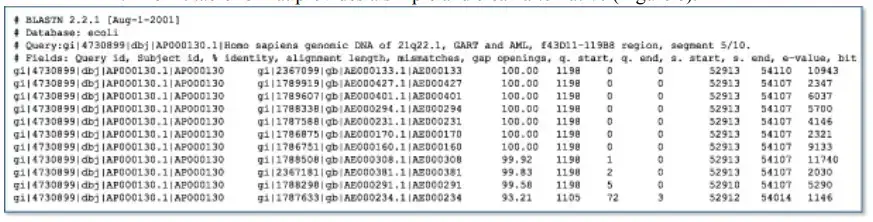
Although the traditional report is optimal for examining the properties of a single gene or protein, scientists frequently wish to perform a large number of BLAST runs for a specific purpose and require only a subset of the information contained in the traditional BLAST report. In cases where the BLAST output will be further processed, it can also be unreliable to parse the conventional report. The traditional report is solely a display format with no formal structure or rules, and it is possible to make improvements at any time by modifying the HTML underneath. The hit table format provides an alternative that is basic and clean. The screening of numerous newly sequenced human Expressed Sequence Tags (ESTs) for contamination by the Escherichia coli cloning vector is an excellent example of when the hit table output is preferable to the traditional report. To differentiate between the contaminating E. coli sequence and the human sequence in this instance, a strict, high E-value threshold would be applied. Human ESTs with extremely robust, near-exact sequence matches with E. coli can be discarded without further examination. (Cases that are borderline may require additional examination by a scientist.) For these purposes, the hit table output is more useful than the standard report because it contains only the required information in a more formal format. The hit table output does not include sequences or definition lines, but it does enumerate, for each matched sequence, the sequence identifier, the start and end points for stretches of sequence similarity (offset by one residue), the percentage identity of the match, and the E-value.
3. Structured Output
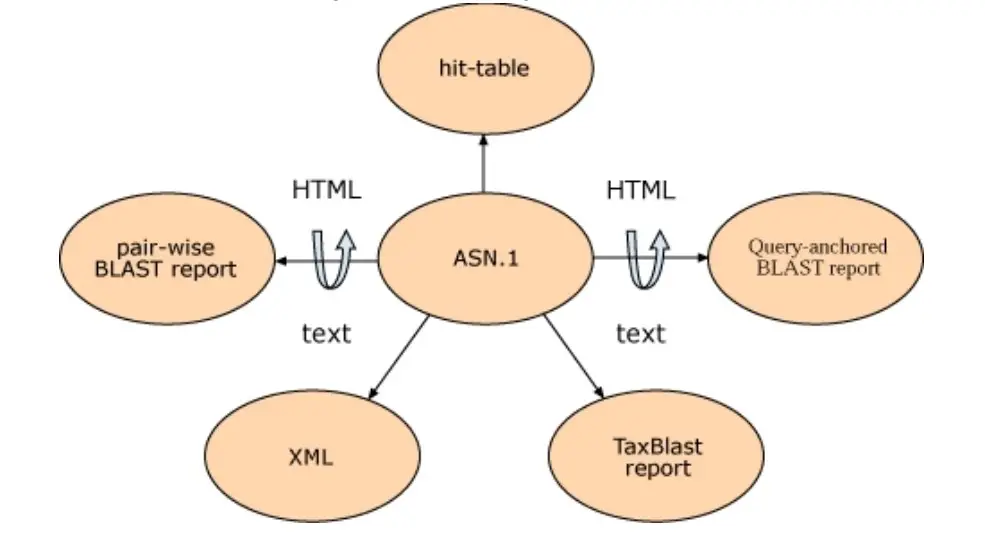
Both parsing the BLAST report and the simpler hit table has disadvantages. It is not possible to automatically check for truncated or otherwise corrupted output when screening a large number of sequences. (This could occur if the disk is filled, for instance.) Additionally, there is no rigorous check for syntax changes in the output, such as the inclusion of new features, which may result in incorrect parsing. Structured output permits rigorous and automatic tests for syntax errors and modifications. XML and ASN.1 are both examples of structured output with built-in syntax and structure validation. (In the case of XML, this is guaranteed by the requirement that tags and the DTD match.) Typically, there is no specification for text reports, but a (incomplete) description of the file is written afterwards.
ASN.1 Is Used by the BLAST Server
In addition to the hit table and traditional HTML report, BLAST results can also be formatted in plain text, XML, and ASN.1 and the format of a particular BLAST result can be altered without rerunning the search. A change in BLAST format is feasible without rerunning the search because, when a scientist views a Web page of BLAST results at NCBI, the HTML for that page is generated from ASN.1. Although the formatted results are requested from the server, the alignment information and corresponding sequences from the BLAST databases are retrieved from a disk in ASN.1 format. The formatter on the BLAST server then compiles these findings into a BLAST report. The BLAST search itself has been decoupled from the format of the result, enabling for multiple output formats from the same search. ASN.1’s stringent internal validation guarantees that these output formats can always be reliably generated.
Information about the Alignment Is Contained within a SeqAlign
SeqAlign is the ASN.1 object that has the BLAST search alignment information. The SeqAlign does not have the actual sequence that was found in the match, but it does have the start, stop, and gap information, as well as scores, E-values, sequence identification, and (DNA) strand information. As was said above, when they are needed, the real database sequences are taken from the BLAST databases. This means that a sequence in the database must have a unique identity. Also, the query sequence can’t have the same number as any other sequence in the database unless the query sequence itself is in the database. If you are using stand-alone BLAST with a custom database, you can use the –O option with formatdb (the tool that changes FASTA files to BLAST database format) to make sure that each sequence has a unique name. This also puts the items in order by their identifiers. In the same way, the –J option in the stand-alone tools blastall, blastpgp, megablast, or rpsblast makes sure that the query does not use an identifier that is already in the database for a different sequence. If the –O and –J options are not used, BLAST gives all sequences unique IDs for that run and hides this information from the user. The uniqueness criterion is already met by any BLAST database or FASTA file from the NCBI website that has gi numbers in it. Unique IDs usually only cause trouble when custom databases are made and the identifiers aren’t given with care. The first token (the letters up to the first space) after the > sign on the defining line is the identifier for a FASTA entry. The easiest case is to have a unique token (like 1, 2, and so on), but you can also make more complicated identifiers that, for example, describe the source of the data. For FASTA identifiers to be parsed accurately, they must follow a certain syntax. Here or in the NCBI Toolkit Software Developer’s guide, you can find more information about the SeqAlign made by BLAST. You can also download a PowerPoint presentation about it.
XML
Both XML and ASN.1 are organized languages that can say the same things. This means that a SeqAlign can be made in XML. Some users don’t like how the information in SeqAlign is set up because it doesn’t show the real sequence, and when it is pulled from the BLAST database, it is packed with two or four bases per byte. Most of the time, these users know what a BLAST report is and want something similar, but in a format that can be safely read. BLAST’s XML file meets this need because it has the query and database sequences, sequence definition lines, the start and end points of the alignments (one offset), and scores, E-values, and percent similarity. For this XML result, there is a public DTD.
How to Use BLAST?
Using BLAST involves a series of steps, from preparing the query sequence to interpreting the results. Here’s a general guide on how to use BLAST:
- Formulate the Query Sequence: Prepare your query sequence that you want to compare against a database. The sequence can be in FASTA format, which starts with a description line followed by the actual sequence.
- Select the BLAST Program: Determine which BLAST program is most appropriate for your sequence type and search requirements. For example, if you have a protein sequence and want to search against a protein database, BLASTp would be suitable.
- Choose the Database: Identify the appropriate database for your search. Common databases include NCBI’s non-redundant (nr) database, protein databases like UniProt, or specific organism-specific databases. The choice depends on your research goals and the sequences you wish to compare against.
- Set Search Parameters: Configure the search parameters based on your needs. This includes selecting the type of algorithm (e.g., BLASTn, BLASTp), specifying the scoring matrix, setting the E-value threshold, and adjusting other parameters such as word size and gap penalties. The default parameters often work well, but adjustments may be needed for specific cases.
- Submit the Search: Submit the query sequence and chosen parameters through a BLAST interface or online tool. Many resources, such as the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov), provide user-friendly interfaces where you can enter your query sequence, select the program and database, and adjust the parameters as needed. Alternatively, you can use command-line versions of BLAST on your local machine.
- Analyze the Results: Once the search is complete, you will receive a list of alignments and statistical measures for each match found. The results typically include information about sequence similarities, alignment scores, E-values, and alignment locations. Evaluate the matches based on the statistical significance, alignment quality, and your research goals.
- Interpret and Refine the Results: Interpret the results in the context of your research question or objectives. Analyze the alignments, identify conserved regions, assess functional annotations, and extract relevant information. You may refine the search parameters, iterate the search with additional queries, or explore further analysis depending on your needs.
Remember to consider the statistical significance of the matches, as indicated by the E-value or other statistical measures. Lower E-values generally indicate more significant matches, but the specific threshold depends on the specific research context.
BLAST is a versatile tool, and the exact steps may vary depending on the BLAST variant, interface, or software used. It is recommended to refer to the documentation or user guides specific to your chosen implementation of BLAST for detailed instructions and guidance.
What is Global and local alignments?
In the context of BLAST (Basic Local Alignment Search Tool), there is a distinction between global alignments and local alignments. BLAST primarily focuses on local alignments. Let’s understand the difference between the two:
- Global Alignment: Global alignment refers to aligning the entire length of two sequences, without considering gaps or mismatches. It attempts to find the best alignment that covers the entire length of both sequences. Global alignment is suitable when comparing sequences that are highly similar or expected to have conserved regions throughout their entire length.
- Local Alignment: Local alignment aims to identify regions of similarity or homology between sequences, even if they are not globally similar. It allows for gaps and mismatches within the alignment to account for sequence variations. Local alignment is particularly useful when comparing sequences that have significant differences but may share short stretches of similarity, such as finding conserved domains or identifying functional motifs within a larger sequence.

BLAST primarily performs local alignments. It starts with a seed search, where it identifies short exact matches (seeds) between the query sequence and the database sequences. Then, it extends these seeds in both directions to find longer regions of similarity, allowing for gaps and mismatches. This process focuses on identifying local regions of significant similarity rather than aligning the entire length of the sequences.
By focusing on local alignments, BLAST efficiently identifies regions of similarity between sequences, even when the sequences as a whole may be dissimilar. This approach is particularly useful for comparing sequences from different organisms, identifying conserved protein domains, detecting functional motifs, and finding similar regions within large genomes or databases.
It’s worth noting that there are other alignment tools, such as Needleman-Wunsch and Smith-Waterman algorithms, which are commonly used for global alignments. However, BLAST’s local alignment approach makes it well-suited for many sequence analysis tasks, given its speed, sensitivity, and ability to handle large databases.
Nucleotides: Word size, and Summary

Proteins: Word size, and Summary

What are Expect values in BLAST?
| E = number of database hits you expect to find by chance, ≥ S |
Expect values, or E-values, are statistical measures that BLAST (Basic Local Alignment Search Tool) uses to figure out how important a match between two sequences is. When a query sequence is compared to a database, the E-value is the predicted number of matches with the same or better scores that would happen by chance.
In BLAST, a match with a smaller E-value is more likely. A small E-value means that the similarity between the query sequence and the database sequence is unlikely to have happened by chance alone and is more likely to be due to homology or a functional link.


The BLAST algorithm’s scoring numbers, such as the alignment scores, the size of the database being searched, and the length of the query sequence, are used to figure out the E-value. It looks at both the size of the database being searched and the size of the query sequence. This gives a measure of the importance that accounts for both the size of the database and the length of the query.
For example, if the E-value of a match is 1e-5 (10-5), it means that, on average, one match with the same or a better score is expected to happen by chance in a collection of the same size for every 105 searches.
It’s important to remember that the E-value should be understood in the context of the research question or study and compared to a threshold that has already been set. The point at which a match is considered significant depends on the area of study and the type of analysis being done. In general, a smaller E-value threshold (like 0.001) means that the criteria for significant matches are more strict.
When using BLAST, it is best to look at the E-value along with other factors like alignment scores, alignment length, and sequence identity to make smart decisions about the biological relevance and importance of the sequence similarity matches.
BLAST Expect Value (In a Nutshell)
- E = number of database hits you expect to find by chance
- As the database size increases …. E increases
- As the score increases …. E decreases
Limits, Errors and Warnings in BLAST
When using the web BLAST search provided by NCBI, there are certain limits, errors, and warnings that you should be aware of. Here are some key points:
- Web BLAST Search Limits:
- Maximum number of target sequences: The web BLAST search has a limit of 5,000 target sequences. This means that if your query matches more than 5,000 sequences in the database, only the top 5,000 matches will be reported.
- Maximum sequence length for nucleotide queries: The web BLAST search allows nucleotide queries up to a maximum length of 1,000,000 bases.
- Maximum sequence length for protein queries: For protein queries, the web BLAST search allows a maximum length of 100,000 amino acids.
It’s important to keep these limits in mind when designing your queries and selecting appropriate databases for comparison. If your query or target sequences exceed these limits, you may need to consider alternative strategies or use other tools that can handle larger data sizes.
- Errors and Warnings:
- Sequence formatting errors: The web BLAST search may generate errors or warnings if there are issues with the formatting of your query sequence. These can include errors related to FASTA format, incorrect characters, or missing sequence information. Make sure to double-check your input sequence for correctness and ensure it meets the required format.
- Database availability: Occasionally, certain databases may not be available or may have limited access due to updates, maintenance, or other technical reasons. If you encounter errors or find a specific database unavailable, you can check the NCBI website or contact NCBI support for more information.
- Resource utilization and waiting time:
- Large queries or databases may require longer processing times. Depending on the size and complexity of your query and database, the BLAST search may take a significant amount of time to complete. It’s important to be patient and allow sufficient time for the analysis to finish.
- Resource limitations: The web BLAST search service is used by a large number of users, and there may be limitations on the number of concurrent jobs or the amount of computational resources available at any given time. If the service is experiencing high demand, you may experience delays or temporary unavailability.


To ensure a smooth experience when using web BLAST, it’s advisable to thoroughly review the provided limits, pay attention to formatting requirements, and be prepared for potential delays or resource limitations. If you require more flexibility or have specific needs that exceed the web BLAST limits, you may consider using a standalone version of BLAST installed locally or exploring alternative bioinformatics tools and resources.
Advantages of BLAST
- Speed and Efficiency: BLAST is designed to deliver fast results, even when searching large sequence databases. It utilizes indexing and heuristic algorithms to accelerate the search process.
- Sensitivity: BLAST is sensitive in detecting remote homologs or sequences with low similarity. It can identify significant matches even in sequences with evolutionary divergence.
- Versatility: BLAST can handle various types of biological sequences, including DNA, RNA, and proteins. It is applicable in a wide range of research areas, from genomics to structural biology.
- User-Friendly Interface: BLAST is accessible to users with different levels of bioinformatics expertise. It provides user-friendly interfaces and online tools that simplify the process of submitting queries and configuring search parameters.
- Community Support and Resources: BLAST has a large and active user community, which has led to extensive resources, tutorials, and documentation. Users can access online forums, documentation, and help from the community to enhance their understanding and usage of BLAST.
Limitations of BLAST
- Sensitivity to Sequence Divergence: BLAST may have difficulty identifying very distantly related sequences due to their low sequence similarity. It may miss subtle functional relationships between sequences.
- False Positives: BLAST may generate false-positive matches due to random similarities or sequence motifs that are not functionally significant. Careful interpretation of results and the use of appropriate statistical thresholds are necessary to minimize false positives.
- Statistical Significance: Determining the statistical significance of BLAST matches can be challenging. The E-value provided by BLAST should be used cautiously and in conjunction with other parameters to assess the significance of matches.
- Alignment Quality: BLAST primarily focuses on local alignments, which may result in alignments with variable quality. Gaps and mismatches are allowed, potentially leading to alignments that are not biologically meaningful.
- Database Bias: BLAST results can be influenced by the composition and representation of the database being searched. Biases in the database can impact the accuracy and interpretation of results, particularly when analyzing sequences from non-model organisms or poorly characterized species.
- Resource Requirements: BLAST searches can be computationally intensive, especially when analyzing large datasets or running complex algorithms. Resource limitations, such as memory and processing power, can impact the speed and scalability of the analysis.
Applications/Uses of BLAST
BLAST (Basic Local Alignment Search Tool) has numerous applications in diverse areas of biological research. Here are some common applications of BLAST:
- Sequence Similarity Search: Comparing a query sequence against a database of known sequences to identify regions of similarity is the primary function of BLAST. It facilitates the discovery of homologous sequences, which can shed light on the functional and evolutionary relationships between genes, proteins, and DNA sequences.
- Genome Annotation: BLAST is an indispensable instrument for genome annotation, which identifies genes and functional elements within a genome. BLAST helps attribute putative functions to newly sequenced genes or identify conserved regions of genomes by comparing genomic sequences to databases of known genes and proteins.
- Prediction of Protein Structure: BLAST can be used to identify structurally related proteins in databases, thereby aiding in protein structure prediction. If a protein with a known structure is identified as a significant match, it can provide valuable information regarding the query protein’s three-dimensional structure.
- Functional Annotation of Genes and Proteins: BLAST can aid in functional annotation by comparing uncharacterized genes or proteins to well-defined sequences. If a query sequence resembles a known sequence with a known function, it provides hints about the query sequence’s prospective function.
- Phylogenetic Analysis: Comparing sequences from various organisms, BLAST facilitates phylogenetic analysis. BLAST serves to reconstruct evolutionary relationships and construct phylogenetic trees by identifying homologous sequences across species.
- Primer Design: BLAST can be utilized to design primers for Polymerase Chain Reaction (PCR) experiments. By aligning primers against a database of sequences, BLAST facilitates the identification of potential binding sites and the evaluation of specificity and cross-reactivity.
- Metagenomic Analysis: In metagenomic research, BLAST is utilized to identify and classify sequences extracted from complex microbial communities. It facilitates the analysis of the diversity and composition of microbial populations, as well as the comprehension of their functional potential.
- Identification of Disease-Related Genes: BLAST identifies disease-related genes by comparing candidate gene sequences with known disease-associated genes. It facilitates the identification of potential disease-causing mutations and the comprehension of the molecular basis of genetic disorders.
- Drug Discovery and Design: BLAST is applicable to drug discovery and design procedures. By comparing the protein or nucleotide sequences of potential drug targets to databases, BLAST identifies potential binding sites, analyzes target homologs, and evaluates the similarity to known drug targets.
- Comparative Genomics: In comparative genomics, BLAST is widely used to compare and analyze the genomes of various organisms. It facilitates the identification of conserved regions, gene families, gene duplications, and genome rearrangements, thereby shedding light on the evolution and function of the genome.
- Development of Diagnostic Tests: BLAST contributes to the development of diagnostic tests for infectious diseases or genetic disorders. BLAST facilitates the identification of specific disease-causing agents or mutations by comparing patient samples to databases of known pathogens or disease-associated variants.
- Functional Genomics: BLAST assists with functional genomics by comparing sequences to functional databases. It contributes to the knowledge of gene function and regulation by aiding in the identification of protein domains, motifs, and functional elements.
- Environmental and Ecological Studies: In environmental and ecological investigations, BLAST is used to analyze microbial communities in various habitats. Metagenomic information can be compared to databases in order to identify and characterize microbial species, their functional potential, and their ecological interactions.
- Vaccine Development: BLAST is involved in the process of vaccine development. It aids in the identification of conserved epitopes across various strains or species, thereby facilitating the development of vaccines that target multiple pathogen variants.
- Evolutionary Studies: In evolutionary studies, BLAST is a valuable tool. It facilitates the comparison of sequences from various species, the identification of orthologs and paralogs, and the reconstruction of evolutionary histories. BLAST is also useful for identifying instances of horizontal gene transfer and researching the evolution of gene families.
- Database Annotation and Validation: Annotation and Validation of Sequence Databases: BLAST is utilized to annotate and validate sequence databases. It aids in the identification of misannotated sequences, the resolution of conflicting annotations, and the improvement of the accuracy and quality of databases.
- Transcriptome Analysis: BLAST is utilized in transcriptome analysis in order to align and compare RNA sequencing (RNA-seq) data against reference databases. It helps with profiling gene expression, analyzing alternative splicing, and identifying novel transcripts.
- MicroRNA Target Prediction: BLAST aids in microRNA (miRNA) target prediction. By comparing miRNA sequences to mRNA databases, BLAST facilitates the identification of potential miRNA target genes.
FAQ
What is BLAST and what is it used for?
BLAST is a bioinformatics tool used for sequence similarity searches. It compares a query sequence against a database to identify similar sequences and infer functional, evolutionary, or structural relationships.
What are the different types of BLAST programs?
The BLAST suite includes various programs for specific sequence types and search requirements, such as BLASTn (nucleotide vs. nucleotide), BLASTp (protein vs. protein), BLASTx (translated nucleotide vs. protein), and tBLASTn (protein vs. translated nucleotide), among others.
How does BLAST determine sequence similarity?
BLAST determines sequence similarity by using algorithms that calculate alignment scores based on matching residues, gaps, and substitution matrices. It identifies local regions of significant similarity rather than performing global alignments.
What is the significance of the E-value in BLAST results?
The E-value (expect value) in BLAST results represents the expected number of matches with similar or better scores that would occur purely by chance. A lower E-value indicates a more significant match.
How do I interpret BLAST results?
BLAST results provide information about sequence alignments, scores, identities, E-values, and more. Interpretation involves assessing statistical significance, alignment quality, and biological relevance, considering factors such as E-values, alignment lengths, and sequence identities.
How do I choose the appropriate BLAST database for my analysis?
The choice of database depends on your research goals and the sequences you want to compare against. Options include public databases like GenBank or UniProt, organism-specific databases, or specialized databases for specific types of sequences (e.g., 16S rRNA).
What are the limitations of BLAST?
BLAST has limitations such as sensitivity to sequence divergence, potential for false positives, and the need for appropriate statistical thresholds. It also has limits on the number of target sequences, sequence lengths, and may have resource limitations in web-based implementations.
Can I use BLAST to analyze my own custom database?
Yes, BLAST allows you to create and search against custom databases. You can prepare a database by formatting your own collection of sequences in a specific format compatible with BLAST.
Are there alternatives to BLAST for sequence similarity searches?
Yes, there are alternative tools for sequence similarity searches, such as FASTA, HMMER, and Smith-Waterman algorithms. Each tool has its own features and strengths, and the choice depends on the specific requirements of your analysis.
Can I run BLAST locally on my computer?
Yes, BLAST is available as standalone software that can be installed on your computer, allowing you to perform local searches. This provides more flexibility and allows you to analyze larger datasets without being limited by web-based implementations.
References
- Wheeler D, Bhagwat M. BLAST QuickStart: Example-Driven Web-Based BLAST Tutorial. In: Bergman NH, editor. Comparative Genomics: Volumes 1 and 2. Totowa (NJ): Humana Press; 2007. Chapter 9. Available from: https://www.ncbi.nlm.nih.gov/books/NBK1734/
- https://bitesizebio.com/21223/how-does-blast-work/
- http://faculty.washington.edu/jht/GS559_2017/lectures/5A-Blast.pdf
- https://www.unmc.edu/bsbc/docs/NCBI_blast.pdf
- https://www.nlm.nih.gov/ncbi/workshops/2022-10_Basic-Web-BLAST/how-blast-works.html
- https://bioinformaticsreview.com/20210503/how-blast-works-concepts-types-methods-explained/