What is Arabinose Operon?
- The L-arabinose operon, also known as the ara or araBAD operon, is essential for the degradation of the five-carbon sugar L-arabinose in Escherichia coli.
- The L-arabinose operon contains three structural genes: araB, araA, and araD (together referred to as araBAD), which code for three metabolic enzymes essential for L-arabinose metabolism.
- These genes generate AraB (ribulokinase), AraA (an isomerase), and AraD (an epimerase), which catalyse the conversion of L-arabinose to D-xylulose-5-phosphate, an intermediate in the pentose phosphate pathway.
- The structural genes of the L-arabinose operon are translated into a single mRNA from a shared promoter.
- The product of regulatory gene araC and the catabolite activator protein (CAP)-cAMP complex govern the expression of the L-arabinose operon as a single unit.
- The regulator protein AraC is arabinose-sensitive and has a dual role as an activator in the presence of arabinose and a repressor in the absence of arabinose to regulate the expression of araBAD.
- At high AraC levels, the AraC protein not only regulates the expression of araBAD, but also regulates its own expression.
- The primary function of the arabinose operon is to convert the complex arabinose molecule into xylulose 5-phosphate, which is then incorporated into the metabolic pathway (pentose phosphate pathway).
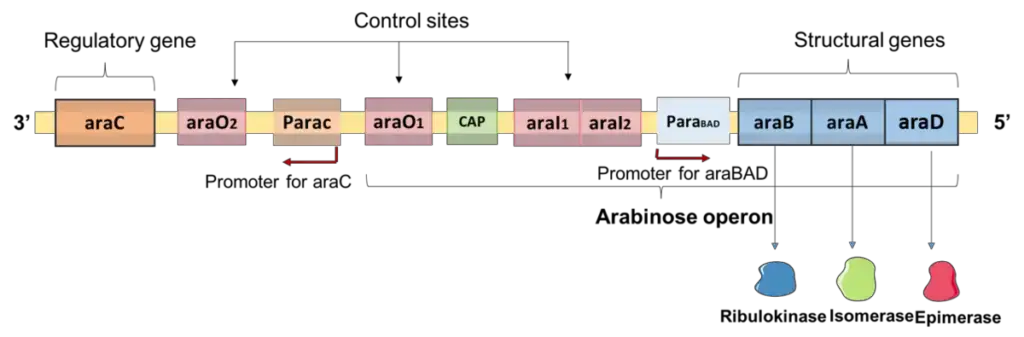
Structure of L-arabinose operon
The linear structure of the arabinose operon consists of four unique genes and a catabolic active site.
- Structural genes
- Inducer genes
- Catabolic active site
- Operator genes
- Promoter genes
- Regulatory gene
Structural genes
- ara-B, ara-A, and ara-D are the three types of structural genes.
- Encoding of metabolic enzymes is the principal function of structural genes.
- Kinase, isomerase, and epimerase are metabolic enzymes that are generated by ara-B, ara-A, and ara-D, respectively.
- The metabolic enzymes degrade the non-glucose molecule, i.e. arabinose, to form a polycistronic or multigenic mRNA.
Function of Structural genes
- araA: L-arabinose isomerase, which catalyses the isomerization of L-arabinose and L-ribulose, is encoded by araA.
- araB: araB encodes ribulokinase, an enzyme that catalyses the phosphorylation of L-ribulose into L-ribulose-5-phosphate.
- araD: L-ribulose-5-phosphate 4-epimerase, which catalyses the epimerization of L-ribulose 5-phosphate and D-xylulose-5-phosphate, is encoded by araD.

Inducer Gene
- Ara I is an inducible gene.
- The transcription is induced by the ara I gene.
- In the arabinose operon, the inducer molecule is arabinose, which binds with the repressor protein to stimulate mRNA transcription.
- The arabinose operon functions positively when an inducer is present and negatively when no inducer is present.
Regulatory gene
- The lone regulatory gene in the arabinose operon is ara-C. The ara-C gene encodes the repressor-functioning Ara C protein.
- Positive and negative regulation of the arabinose operon by the Ara C protein.
- When the Ara C protein attaches to the operator, it inhibits araBAD mRNA synthesis. It will not increase RNA polymerase binding to the promoter region.
- The complex is activated when the Ara C repressor protein connects with the inducer, arabinose. This activation of Ara C with arabinose leads to the synthesis of araBAD mRNA by promoting the attachment of RNA polymerase to the promoter region.
Catabolic Active Site
- CAP is the activator site of the arabinose operon.
- CAP stands for catabolite activator protein, which promotes the efficient binding of the RNA polymerase to the promoter region to increase the efficiency of transcription rate.
- When glucose is abundant and arabinose is scarce, ATP cannot be converted into cAMP.
- If glucose availability is low and arabinose content is high, ATP will convert to cAMP, and the whole complex will bind to the CAP to trigger mRNA transcription.
- The complex formed by cyclic AMP and CAP will bind to the CAP region of the operon.
Operator Gene
- Two operator genes, designated O1 and O2, exist (operate araBAD mRNA synthesis).
- In positive regulation, when the amount of glucose is low and the amount of arabinose is high, the arabinose will activate the repressor protein (Ara C). It will stimulate the production of araBAD mRNA.
- In negative regulation, when glucose levels are high and arabinose levels are low, the repressor protein (Ara C) will not induce the production of araBAD mRNA.
Promoter Gene
- In the arabinose operon, there are two promoter genes, PBAD and PC.
- PBAD is the promoter site of structural ara BAD genes, promoting the synthesis of ara BAD mRNA.
- PC is the promoter region of the ara C regulatory gene, which promotes the manufacture of the Ara C repressor protein.
- In the absence of inducer, the Ara C repressor exists in the active P1 state, while in the inactive P2 state (in the presence of inducer).
In Summary
- L-arabinose operon consists of structural genes and regulatory sections, such as the operator region (araO1, araO2) and the initiator region (araI1, araI2).
- araB, araA, and araD are structural genes that encode enzymes for L-arabinose catabolism.
- There is also a CAP binding site to which the CAP-cAMP complex attaches and aids catabolite suppression, resulting in the positive regulation of araBAD when the cell is glucose-starved.
- Located upstream of the L-arabinose operon, the regulatory gene araC encodes the arabinose-responsive regulatory protein AraC.
- Both araC and araBAD contain a promoter where RNA polymerase interacts and transcription is initiated.
- araBAD and araC are transcribed in opposite directions from their respective promoters, araBAD (PBAD) and araC (PC).
Regulation of Arabinose Operon
- The L-arabinose system is not only controlled by the CAP-cAMP activator, but also favourably or negatively regulated by the AraC protein.
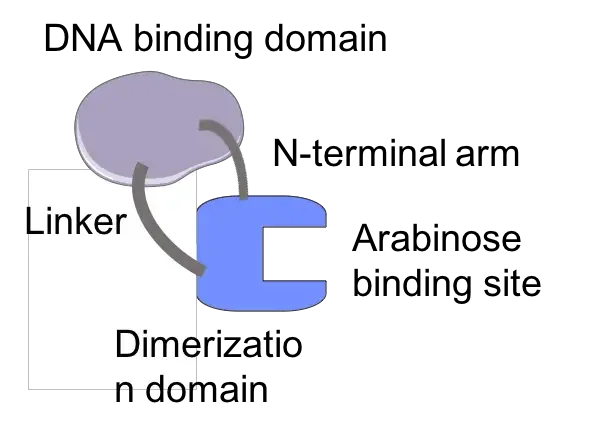
- AraC is a homodimer that regulates transcription of araBAD by interacting with the operator and initiator area of the L-arabinose operon.
- Each AraC monomer contains two domains: a DNA-binding domain and a dimerisation domain.
- The dimerisation domain is in charge of binding arabinose. Upon arabinose binding, AraC undergoes a conformational shift, resulting in two different conformations.
- The conformation is dictated exclusively by the binding of the allosteric inducer arabinose.
- When the concentration of AraC is too high, AraC is also able to negatively autoregulate its own expression.
- AraC synthesis is inhibited by dimeric AraC binding to the operator region (araO1).
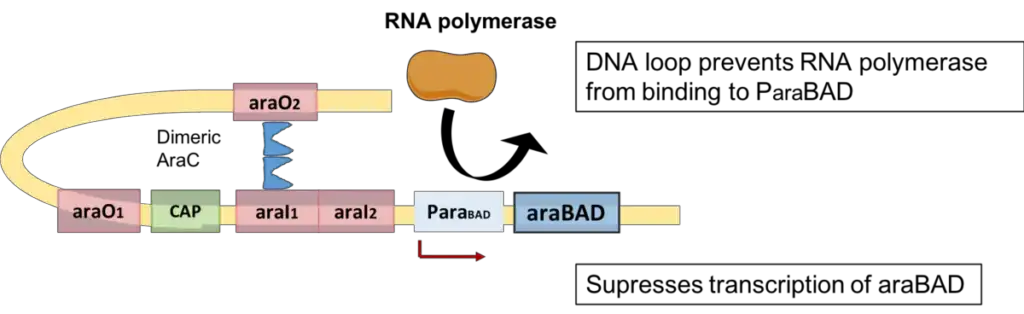
Negative regulation of araBAD
- Absent arabinose, cells do not require araBAD products for arabinose degradation.
- Consequently, dimeric AraC functions as a repressor: one monomer binds to the operator of the araBAD gene (araO2), and the other monomer attaches to a remote DNA half site known as araI1.
- This ultimately results in the development of a DNA loop. This orientation inhibits the binding of RNA polymerase to the araBAD promoter.
- Consequently, araBAD structural gene transcription is suppressed.
Positive regulation of araBAD
In the absence of glucose and in the presence of arabinose, the araBAD operon is active. When arabinose is present, both AraC and CAP serve as activators in conjunction.
Role of AraC
- AraC works as an activator when arabinose is present.
- When arabinose binds to the AraC dimerization domain, AraC experiences a conformational shift. Consequently, the AraC-arabinose complex dissociates from araO2 and break the DNA loop.
- Therefore, in the presence of arabinose, it is more energetically favourable for AraC-arabinose to bind to two nearby DNA half sites: araI1 and araI2.
- One of the monomers binds araI1, while the other binds araI2; hence, arabinose induces allosterically the binding of AraC to araI2.
- In this structure, one of the AraC monomers positions close to the araBAD promoter, which helps to recruit RNA polymerase to the promoter to commence transcription.
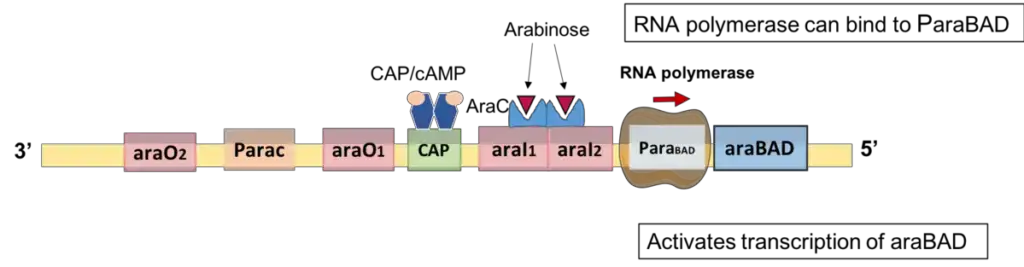
Role of CAP/cAMP (catabolite repression)
- CAP only acts as a transcriptional activator when glucose, E. coli’s preferred sugar, is absent.
- In the absence of glucose, a high concentration of CAP protein/cAMP complex binds to the CAP binding site, which is located between araI1 and araO1.
- The binding of CAP/cAMP is responsible for opening the DNA loop between araI1 and araO2, increasing the binding affinity of AraC protein for araI2, and promoting RNA polymerase to bind to the araBAD promoter in order to activate the expression of the araBAD gene required for metabolising L-arabinose.
Autoregulation of AraC
- AraC, the protein product of araC, negatively regulates its own expression.
- At high levels of AraC, excess AraC binds to the operator of the araC gene, araO1, preventing RNA polymerase from accessing the araC promoter.
- Therefore, at high doses, the AraC protein blocks its own expression.
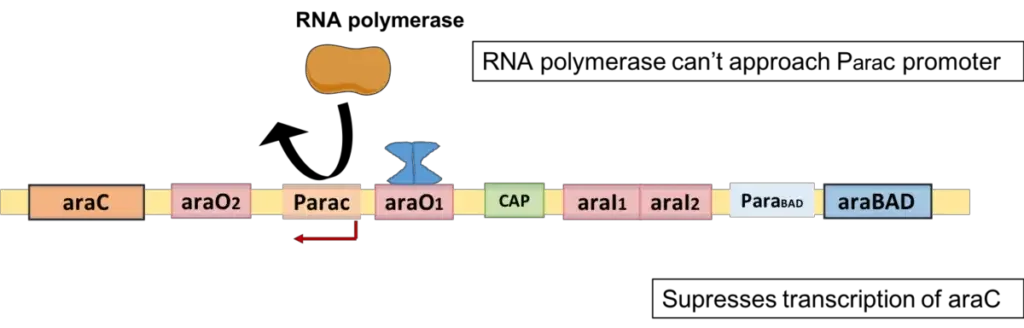
Effect of Arabinose Operon on mRNA Synthesis
Similar to the Lac operon, the arabinose operon is both positively and negatively regulated.
During Positive Regulation of Arabinose Operon
A positive phrase here suggests that mRNA synthesis will occur. Consequently, ara BAD mRNA is produced under positive regulation. Arabinose operon is positively regulated by the two aforementioned situations.
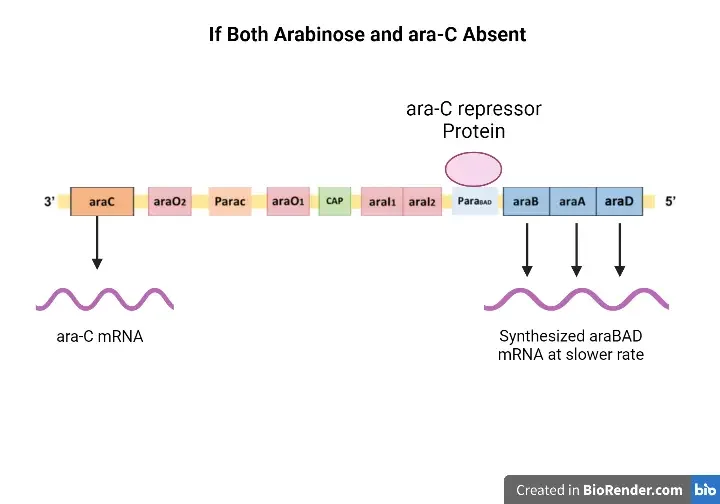
When both inducer and repressor protein is absent
- The arabinose operon will not be repressed in the absence of both an inducer and a repressor protein.
- RNA polymerase will attach to the particular promoter region and transcribe the ara-BAD genes into mRNA under this situation. In this instance, however, the rate of mRNA transcription is significantly slower.
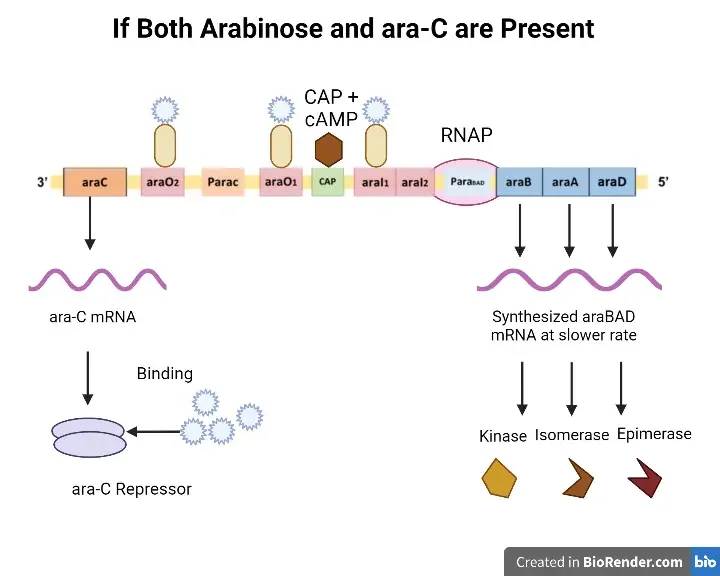
When both inducer and repressor are present
- To regulate mRNA transcription, the inducer (arabinose) will bind to the repressor protein.
- Ara-C protein and arabinose produce a compound that prevents the creation of a loop.
- Arabinose interacts with the Ara-C dimer to alter its structural conformation.
- This modification to the structural arrangement will permit the RNA polymerase to transcribe the ara-BAD genes into mRNA.
- The mRNA will eventually be converted into proteins.
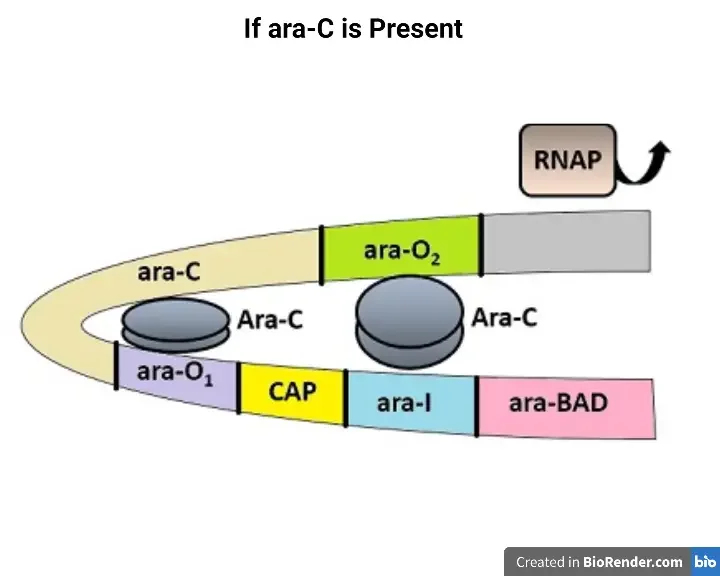
During Negative Regulation of Arabinose Operon
A negative word means that mRNA transcription will not take place. To simplify the explanation, let’s compare the third condition to the previous two.
When only repressor protein is present
- The primary function of the arabinose operon is the degradation of arabinose.
- There will be no transcription and translation of the DNA molecule in the absence of arabinose.
- Arabinose functions as an inducer, binding and inactivating the ARA-C repressor protein.
- In the absence of inducer, the ara-C gene will generate the repressor protein. By forming a loop, the ARA-C repressor protein forms a dimer with the operator and the inducer gene.
- The creation of the loop will prevent the RNA polymerase from transcribing the ara-BAD genes into mRNA.
Effect Of Mutations On Ara Genes
Point mutations in the genetic sequences of any of the ara genes can have profound effects on bacterial metabolism.
- Mutations in the ara-C gene (which controls the expression of the 3 structural genes) result in a super-repressed state in which ara-A, ara-B, and ara-D (3 structural genes) are shut down even in the presence of copious arabinose, i.e. arabinose operon is shut down.
- A mutation in the ara-A gene will render the bacterium arabinose-negative, unable to utilise arabinose as a carbon source.
- Additionally, a mutation in the ara-B gene will render the bacterium arabinose-negative.
- Due to the accumulation of ribulose 5 phosphate, which is poisonous to cells in large concentrations, a mutation in the ara-D gene will result in cell death.
References
- Schleif, R. (2010). AraC protein, regulation of the l-arabinose operon inEscherichia coli, and the light switch mechanism of AraC action. FEMS Microbiology Reviews, 34(5), 779–796. doi:10.1111/j.1574-6976.2010.00226.x
- Schleif, R. F. (2004). ara Operon. Encyclopedia of Biological Chemistry, 116–119. doi:10.1016/b0-12-443710-9/00239-8
- Schleif, R. (2000). Regulation of the ?-arabinose operon of Escherichia coli. Trends in Genetics, 16(12), 559–565. doi:10.1016/s0168-9525(00)02153-3
- Englesberg E, Squires C, Meronk F Jr. The L-arabinose operon in Escherichia coli B-r: a genetic demonstration of two functional states of the product of a regulator gene. Proc Natl Acad Sci U S A. 1969 Apr;62(4):1100-7. doi: 10.1073/pnas.62.4.1100. PMID: 4894687; PMCID: PMC223620.
- (2008). Arabinose Operon. In: Encyclopedia of Genetics, Genomics, Proteomics and Informatics. Springer, Dordrecht. https://doi.org/10.1007/978-1-4020-6754-9_1090
- Arabinose Operon. (2008). Encyclopedia of Genetics, Genomics, Proteomics and Informatics, 137–138. doi:10.1007/978-1-4020-6754-9_1090
- Englesberg, E., Squires, C., & Meronk, F. (1969). THE L-ARABINOSE OPERON IN Escherichia coli B/r: A GENETIC DEMONSTRATION OF TWO FUNCTIONAL STATES OF THE PRODUCT OF A REGULATOR GENE. Proceedings of the National Academy of Sciences, 62(4), 1100–1107. doi:10.1073/pnas.62.4.1100
- Rodgers, Michael & Schleif, Robert. (2008). DNA tape measurements of AraC. Nucleic acids research. 36. 404-10. 10.1093/nar/gkm1030.
- https://grantome.com/grant/NIH/R37-GM018277-29
- https://www.sas.upenn.edu/labmanuals/biol123/Table_of_Contents_files/Appendix-AraOperon.pdf
- https://www.slideshare.net/SyedMuhammadKhan/arabinose-operon
- https://biologyreader.com/arabinose-operon.html
- https://en.wikipedia.org/wiki/L-arabinose_operon
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.