What is Whole Genome Sequencing?
- Whole genome sequencing (WGS) is a method used to determine the complete DNA sequence of an organism’s genome in a single effort. This includes sequencing all of the organism’s chromosomal DNA, as well as the DNA found in mitochondria, and in plants, chloroplasts.
- The process of whole genome sequencing provides a comprehensive view of the entire genome, unlike other sequencing methods that target specific regions, such as exome sequencing. WGS is crucial in research and has begun to be implemented in clinical settings. It’s anticipated to play a significant role in personalized medicine by guiding therapeutic interventions based on an individual’s genomic data.
- Historically, DNA sequencing methods were manual and labor-intensive. For instance, the Maxam-Gilbert and Sanger sequencing methods, developed in the 1970s and 1980s, required meticulous manual procedures. These early techniques were used to sequence several whole bacteriophage and animal viral genomes. However, the advent of automated sequencing methods in the 1990s marked a significant advancement, enabling the sequencing of larger bacterial and eukaryotic genomes more efficiently.
- The first organism to have its complete genome sequenced was the bacteriophage MS2 in 1976. In 1992, the yeast chromosome III was the first chromosome of any organism to be fully sequenced. Haemophilus influenzae, in 1995, was the first organism to have its entire genome sequenced. H. influenzae has a genome of approximately 1.8 million base pairs. In contrast, the genomes of eukaryotic organisms, such as the amoeba Amoeba dubia and humans, are much larger. For example, A. dubia has a genome size of around 700 billion nucleotide pairs, while the human genome contains about 3.2 billion nucleotide pairs.
- The sequencing of eukaryotic genomes has utilized various methods. Shotgun sequencing, a method where DNA is fragmented and then reassembled, was pivotal in sequencing early bacterial and archaeal genomes. This method was also used to sequence the first eukaryotic genome, that of Saccharomyces cerevisiae (yeast), in 1996. S. cerevisiae’s genome comprises approximately 12 million nucleotide pairs. The first multicellular eukaryote and animal genome sequenced was that of the nematode Caenorhabditis elegans in 1998.
- Significant milestones in genome sequencing include the publication of the entire DNA sequence of human chromosome 22 in 1999 and the sequencing of the fruit fly Drosophila melanogaster genome in 2000. The first plant genome, that of Arabidopsis thaliana, was also fully sequenced by 2000. A draft of the human genome was published in 2001, and the genome of the laboratory mouse Mus musculus was completed in 2002. The Human Genome Project published an incomplete version of the human genome in 2004, and in 2008, the first female human genome was sequenced.
- Today, thousands of genomes have been wholly or partially sequenced, with advancements in next-generation sequencing (NGS) making the process faster and more affordable. This technological progress has transformed WGS into a practical tool for both research and clinical applications, paving the way for advancements in our understanding of genetics and the development of personalized medicine.
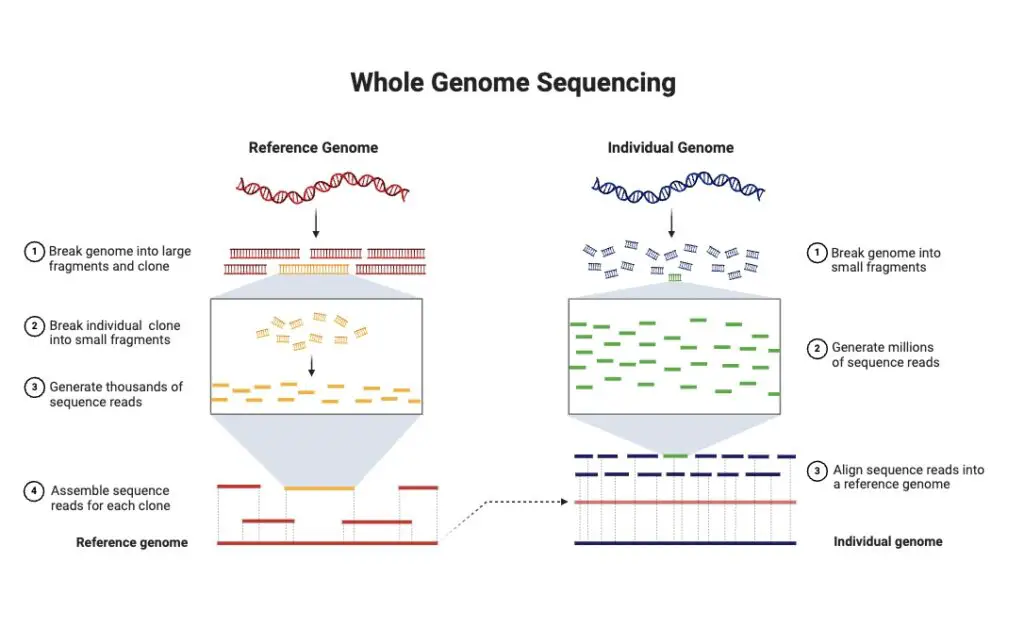
Principle of Whole Genome Sequencing
The principle of whole genome sequencing (WGS) revolves around the complete sequencing of an organism’s DNA, encompassing both coding and non-coding regions. This method provides a thorough understanding of the genome, detailing the genes, regulatory elements, and variations present.
WGS begins with the extraction of DNA from the organism. The extracted DNA is then used to construct a sequencing library. This involves fragmenting the DNA into smaller pieces, which are then sequenced. The sequence data obtained from these fragments is analyzed to identify genetic variations and to reconstruct the entire genome.
WGS employs two main strategies: shotgun sequencing and pairwise-end sequencing. In shotgun sequencing, the DNA is randomly fragmented into smaller pieces. Each fragment is sequenced individually. The sequences are then analyzed to find overlaps, which helps in assembling the entire genome sequence. This method is effective for sequencing large genomes, as it allows for the parallel sequencing of many fragments.
Pairwise-end sequencing, on the other hand, involves sequencing both ends of each DNA fragment. This method provides more contextual information about the location of the fragments, aiding in the accurate reconstruction of the genome. By knowing the sequences at both ends, scientists can determine how the fragments overlap, which enhances the accuracy of the genome assembly.
The detailed process of WGS includes several key steps. Firstly, the extraction of high-quality DNA is crucial, as it ensures the reliability of the sequencing data. Next, the construction of the sequencing library involves the preparation of DNA fragments suitable for sequencing. This is followed by the actual sequencing of the DNA fragments, where advanced sequencing technologies are employed to read the nucleotide sequences.
After sequencing, bioinformatic tools are used to analyze the sequence data. These tools help in assembling the fragments into a continuous sequence, identifying genetic variations, and annotating genes and regulatory elements. The final genome sequence provides comprehensive insights into the organism’s genetic makeup, which can be used for various applications in research and medicine.
Therefore, the principle of whole genome sequencing is centered on obtaining a complete and detailed view of an organism’s genome. By sequencing the entire DNA, including non-coding regions, WGS offers a holistic understanding of genetic information, facilitating advances in genomics and personalized medicine.
Types of Whole Genome Sequencing
Whole genome sequencing (WGS) can be divided into two main types: de novo genome sequencing and whole-genome resequencing (WGR). Each type serves distinct purposes and involves different methodologies.
1. De Novo Genome Sequencing
De novo genome sequencing is used to assemble new genomes that lack a prior reference sequence. This approach is essential for sequencing newly studied species or genomes that exhibit high variability. The term “de novo” means “from the beginning,” reflecting the process of constructing a genome sequence from scratch.
Purpose and Applications:
- New Species: This method is invaluable for creating reference genomes for new species. By providing foundational sequence data, de novo sequencing enables further genomic studies.
- Genomic Diversity: It is used to explore genomes with significant variability, offering insights into unique genetic characteristics.
Process:
- DNA Fragmentation: The genome is fragmented into smaller pieces.
- Sequencing: Each fragment is sequenced individually.
- Assembly: Advanced bioinformatic tools are used to assemble the sequences, reconstructing the entire genome without any reference.
Challenges:
- Complexity: The process can be demanding due to the genome’s complexity.
- Bioinformatics: Extensive resources and expertise are required for accurate assembly and analysis.
2. Whole-Genome Resequencing (WGR)
Whole-genome resequencing (WGR) involves sequencing the genome of an individual or population and comparing it to an existing reference genome. This method focuses on identifying genetic variants by mapping sequence reads to the reference genome.
Purpose and Applications:
- Variant Identification: WGR is widely used to pinpoint genetic variants, such as single nucleotide polymorphisms (SNPs) and structural variations.
- Genetic Diversity: It helps study genetic diversity within populations, providing insights into evolutionary biology and disease susceptibility.
Process:
- Sequencing: The genome of an individual or population is sequenced.
- Read Mapping: The obtained sequence reads are mapped to an existing reference genome.
- Variant Analysis: Differences between the sequenced genome and the reference genome are identified and analyzed.
Advantages:
- Efficiency: WGR is more efficient than de novo sequencing as it leverages an existing reference genome.
- Precision: The use of a reference genome enhances the accuracy of variant detection.
Limitations:
- Dependence on Reference: The accuracy of WGR is contingent upon the quality and completeness of the reference genome.
- Limited Novel Discovery: Since it relies on an existing reference, WGR may not be as effective in discovering completely new genetic elements.
Steps of Whole Genome Sequencing (WGS)
Whole Genome Sequencing (WGS) is a comprehensive method used to determine the complete DNA sequence of an organism’s genome. The process involves several critical steps that ensure the accurate and detailed sequencing of the genome.
1. Sample Preparation
The initial step in WGS is sample preparation, which involves obtaining high-quality nucleic acid samples.
- Sample Collection: Biological samples are collected from the organism of interest.
- Cell Lysis: Cells are lysed using physical or chemical methods to release DNA.
- DNA Purification: The DNA is purified from proteins, lipids, and other cellular debris using various extraction methods.
2. Library Construction
After purifying the nucleic acids, the next step is constructing a sequencing library, which includes short DNA fragments.
- DNA Fragmentation: The genomic material is fragmented into required lengths using mechanical shearing or enzymatic digestion.
- End Repair and Adapter Ligation: The fragmented DNA undergoes end repair, followed by the ligation of adapters to the ends. These adapters contain sequences essential for sequencing.
- Library Enrichment: The adapter-ligated library is enriched to ensure a high concentration of DNA fragments.
- Quality Validation: The constructed library is validated for quality to meet the requirements of sequencing instruments.
3. Sequencing
The prepared library is then sequenced using a chosen sequencing platform.
- Loading: The library is loaded onto the sequencing platform.
- Sequencing Technologies: Next-generation sequencing (NGS) platforms such as Illumina, PacBio, and Oxford Nanopore are commonly used. These platforms can generate massive quantities of short reads with varying lengths.
- Data Formatting: The sequencing output is formatted into standardized files for subsequent alignment and analysis.
4. Alignment and Assembly
Alignment maps the short nucleotide reads to a reference genome, while assembly reconstructs the genome sequences into larger contiguous segments.
- Alignment: Short reads are mapped to a reference genome using tools like BOWTIE, BWA, and SOAP2. This step is computationally intensive due to the vast number of possible positions in a reference genome.
- Assembly Methods:
- Reference-Based Assembly: Aligns reads to an existing reference genome, producing a sequence that closely matches the reference. It requires fewer computational resources but cannot generate novel sequences.
- De Novo Assembly: Uses computational methods to align overlapping reads without relying on a reference genome. This method is crucial for discovering new sequences but requires significant computational resources. Tools include Velvet, SOAPdenovo, and ABySS.
5. Quality Control
Quality control ensures that sequencing errors are minimized, leading to more accurate biological analyses.
- Data Checking: Raw sequencing data is checked for quality metrics such as read length, primer contamination, and adapter contamination.
- Error Identification: Low-quality reads and those containing adapters are identified and removed.
- Quality Metrics: Metrics such as N50, assembly size, and contig numbers assess the quality of assembled genomes. Tools like FastQC and PRINSEQ are commonly used.
6. Variant Calling
Variant calling identifies differences between the sequenced genome and the reference genome to detect genetic variants.
- Variant Detection: Tools categorize variants as single-nucleotide polymorphisms (SNPs), insertions or deletions (indels), structural variants (SVs), and copy number variations (CNVs).
- Common Tools: GATK, SAMtools, and SOAPsnp are used for variant calling.
7. Annotation
Genome annotation adds biological information to the sequenced data and identified variants.
- Structural Annotation: Predicts the locations and structural components of genes and other genomic elements. Tools like AUGUSTUS and GeneMark identify open reading frames (ORFs).
- Functional Annotation: Assigns functions to the predicted genes by comparing them to existing databases. BLAST and InterProScan are commonly used.
- Variant Annotation: Tools such as ANNOVAR and VEP annotate the identified variants.
8. Analysis
The final step is interpreting and analyzing the annotated data to translate sequencing data into meaningful biological insights.
- Pathway Analysis: Identifies biological pathways and the functional impact of variants using databases like KEGG and Reactome.
- Population Genetics: Analyzes genetic diversity and provides information about evolutionary history and genetic risk factors for diseases.
- Comparative Genomics: Compares genomes and constructs phylogenetic trees using tools like MEGA and PhyML.
- Gene Expression Analysis: Uses RNA-seq data to study gene expression patterns, while epigenetic analysis examines DNA modifications like methylation.
Advantages of Whole Genome Sequencing
- Rapid Identification and Characterization of Microorganisms
WGS facilitates the quick identification of microorganisms by providing a complete genetic blueprint. This method allows for the determination of strain relatedness, geographical origin, and evolutionary history. Consequently, it aids in understanding microbial diversity and tracking the spread of pathogens. - Identification of Critical Virulence Factors
Through WGS, researchers can pinpoint unique virulence factors that enable pathogens to cause diseases. These factors are crucial for understanding how microorganisms interact with hosts and contribute to illness, guiding the development of targeted treatments and preventive measures. - Detection of Antimicrobial Resistance
WGS can rapidly identify antimicrobial resistance profiles of microorganisms. By analyzing the entire genome, it detects which antibiotics are ineffective against specific strains, providing faster and more comprehensive data than traditional culture methods. This information is vital for effective treatment planning and managing resistance. - High-Resolution Genomic Representation
WGS offers a detailed, base-by-base view of the genome. This high-resolution representation captures both large-scale and small genetic variants that other techniques may miss. Such thoroughness is essential for accurate genetic analysis and understanding complex genomic variations. - Detection of Genetic Variants
WGS uncovers a broad spectrum of genetic variations, including single nucleotide variations (SNVs) and small insertions or deletions (indels). It provides high accuracy in detecting these variations, which is critical for precise genetic analysis and research. - Insight into Gene Expression and Regulatory Mechanisms
WGS identifies variants in both protein-coding and non-coding regions of the genome. This comprehensive approach provides valuable information about gene expression and regulatory mechanisms. It helps in understanding how genetic changes influence gene function and regulation. - Efficient Data Delivery
WGS generates large volumes of data rapidly, facilitating the assembly of new genomes and genetic analyses. The quick turnaround of data supports timely research and the development of new genetic insights. - Outbreak Detection and Analysis
WGS is instrumental in the detection, mapping, and analysis of outbreaks. It enables the swift identification and tracking of pathogens, which is crucial for controlling and mitigating the spread of infectious diseases.
Limitations of Whole Genome Sequencing
- Variants of Uncertain Significance
WGS frequently identifies numerous genetic variants whose significance is unclear. The sheer volume of data can make it challenging to determine the clinical relevance of these variants. Consequently, this uncertainty can complicate the interpretation of results and their implications for disease diagnosis and treatment. - Challenges in Analyzing Repetitive Regions
Certain regions of the genome, particularly those with repetitive sequences, may not be accurately analyzed with WGS. This limitation can lead to gaps in genomic data and hinder the complete understanding of genomic structures and variations in these regions. - High Costs
Despite advancements that have reduced the cost of WGS, it remains expensive, especially for large-scale studies or clinical applications. The financial burden can limit access to WGS and its widespread adoption in certain contexts. - Need for Powerful Computational Resources
The large volume of data generated by WGS necessitates substantial computational resources for effective data processing and analysis. This requirement can be a barrier for institutions with limited computational infrastructure and expertise. - Ethical and Privacy Concerns
The extensive genetic data produced by WGS raises significant ethical issues. Concerns include privacy, informed consent, and the potential misuse of genetic information. Ensuring the protection of individuals’ genetic data and addressing these ethical considerations are crucial for responsible WGS practices.
Applications of Whole Genome Sequencing
Whole Genome Sequencing (WGS) is a powerful tool with diverse applications in genomics and medicine. It provides comprehensive insights into the genetic makeup of organisms, offering a broad range of uses in various fields. The following are key applications of WGS:
- Identification of Novel Disease Genes
WGS enables the discovery of previously unknown genes linked to diseases. By analyzing the entire genome, researchers can uncover new genetic variations associated with different conditions. This expands the understanding of disease mechanisms and identifies potential targets for therapeutic intervention. - Population Genetics and Evolutionary Studies
WGS facilitates the study of genetic diversity within and between populations. This application is crucial for understanding human evolution and migration patterns. By comparing genomes from different populations, scientists can trace historical migrations and identify genetic adaptations to environmental changes. - Functional Genomics
Through WGS, researchers can investigate the impact of genetic mutations on protein function. This application helps in understanding how changes in DNA affect protein structure and function, which is vital for developing new therapies and drug targets. It provides insights into the functional consequences of genetic variations. - Epigenetic Research
Combining WGS with epigenetic profiling allows for a comprehensive analysis of gene regulation. This approach explores how epigenetic modifications, such as DNA methylation and histone modification, influence gene expression and contribute to disease development. It helps elucidate the interplay between genetics and epigenetics. - Developmental Biology
WGS is instrumental in studying genetic factors underlying developmental disorders and birth defects. By analyzing the genomes of individuals with developmental anomalies, researchers can gain insights into the molecular mechanisms that drive normal and abnormal development. This understanding can lead to better diagnostic and therapeutic strategies. - Cancer Diagnosis and Treatment
In oncology, WGS is used to identify somatic mutations in cancer cells. This allows for a more accurate diagnosis and the development of personalized treatment plans. By pinpointing specific mutations, clinicians can tailor therapies to target the unique genetic profile of each tumor, improving treatment outcomes. - Pharmacogenomics
WGS helps in understanding how genetic variations affect drug metabolism, efficacy, and toxicity. This application is crucial for personalized medicine, as it enables healthcare providers to select the most effective and safest medications based on an individual’s genetic makeup. It enhances the precision of drug prescribing and minimizes adverse effects.
References
- Goodwin, S., McPherson, J. D., & McCombie, W. R. (2016). Coming of age: ten years of next-generation sequencing technologies. Nature Reviews Genetics, 17(6), 333-351.
- Metzker, M. L. (2010). Sequencing technologies—the next generation. Nature Reviews Genetics, 11(1), 31-46.
- Fuentes-Pardo, A. P., & Ruzzante, D. E. (2017). Whole-genome sequencing approaches for conservation biology: Advantages, limitations and practical recommendations. Molecular Ecology, 26(20), 5369–5406. https://onlinelibrary.wiley.com/doi/10.1111/mec.14264
- Besser, J., Carleton, H. A., Gerner-Smidt, P., Lindsey, R. L., & Trees, E. (2018). Next-generation sequencing technologies and their application to the study and control of bacterial infections. Clinical Microbiology and Infection, 24(4), 335-341.
- Schwarze, K., Buchanan, J., Taylor, J. C., & Wordsworth, S. (2018). Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genetics in Medicine, 20(10), 1122-1130.
- Ekblom, R., & Wolf, J. B. (2014). A field guide to whole-genome sequencing, assembly and annotation. Evolutionary Applications, 7(9), 1026–1042. https://doi.org/10.1111/eva.12178
- Gullapalli, R. R., Desai, K. V., Santana-Santos, L., Kant, J. A., & Becich, M. J. (2012). Next generation sequencing in clinical medicine: Challenges and lessons for pathology and biomedical informatics. Journal of Pathology Informatics, 3, 40.
- Burian, A. N., Zhao, W., Lo, T. W., & Thurtle-Schmidt, D. M. (2021). Genome sequencing guide: An introductory toolbox to whole-genome analysis methods. Biochemistry and molecular biology education: a bimonthly publication of the International Union of Biochemistry and Molecular Biology, 49(5), 815–825. https://doi.org/10.1002/bmb.21561
- Yin, R., Kwoh, C.K. and Zheng, J., 2019. Whole genome sequencing analysis.
- Uelze, L., Grützke, J., Borowiak, M. et al. Typing methods based on whole genome sequencing data. One Health Outlook 2, 3 (2020).
- Brunfeldt, M., Teare, H., Schuurbiers, D. et al. Simulating the Genetics Clinic of the Future — whether undergoing whole-genome sequencing shapes professional attitudes. J Community Genet 13, 247–256 (2022)
- Amor, D.J., 2015. Future of whole genome sequencing. Journal of Paediatrics and Child Health, 51(3), pp.251-254.
- https://sciencevivid.com/whole-genome-sequencing-wgs-introduction-workflow-pipelines-applications/