What is Transposition?
Transposition is the process in which a particular DNA sequence is shifted from one position of the genome to another position. It is the movement of a transposable element or jumping gene, and it is not dependent on any homologous sequence at the target site. It is the process that may occur in the same chromosome or it may occur between different chromosomes.
The DNA fragment that participate in this step is referred to as a transposon, and it is inserted into a new location while altering the earlier genetic arrangement. It is the process that may follow either a cut-and-paste type mechanism where the element is excised from its original site or a copy-and-paste type mechanism where an RNA intermediate is formed before insertion.
It is the reason for various changes in the genome because insertion of a transposon may cause mutation and sometimes changes in gene expression.
Among the important findings in genetics, the discovery of transposition was first made by Barbara McClintock while she studied maize, and these elements are now known to be present in almost all organisms including bacteria and humans.
Types and Mechanisms of Transposition
- Transposons are commonly referred to as “jumping genes” because of their capacity to jump from one location to another.
- However, the word is rather deceptive as it indicates that DNA always leaves one location and moves to another.
- This type of transposition is known as nonreplicative transposition or “cut and paste” because both strands of the original DNA transfer from one location to another without reproducing.
- Nevertheless, transposition typically involves DNA replication, so one copy of the transposon remains at its original location while another copy inserts at the new location. This is known as replicative transposition (or “copy and paste”) because a transposon that moves along this route repeats itself.
- Consequently, there are two forms of Transposition, including:
- nonreplicative transposition (or “cut and paste”)
- replicative transposition (or “copy and paste”)
1. Replicative Transposition
- When a transposon replicates, making a new copy while leaving the old copy behind, it is referred to as a replicative transposon, however when transposons travel from one location to another, leaving a gap behind, they are referred to as non-replicative transposons.
- Simply said, replicative and non-replicative transposition are also referred to as copy-and-paste and cut-and-paste transposition, respectively.
Replicative transposition of DNA transposons
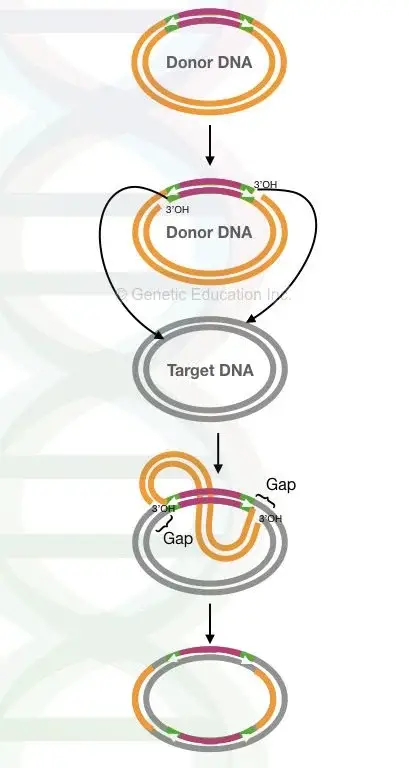
- At the beginning of the process, the transposase enzyme is assembled at both ends of the transposon. It is the step where the whole element is prepared for movement. The assembly makes the active transposon and it is the process helping in cutting at one place and joining at another place. This is referred to as the cleaving and joining reaction.
- The transposase protein now acts on both ends of the element and creates free 3′ OH ends. It is the step where a space is made in the target DNA. These enzyme also nick the donor ends and the target site, so 4 nicks is formed in total. The nicks on the transposon ends have same sequences because of inverted terminal repeats. The major thing here is production of free 3′ ends which is important for polymerase to fill gaps.
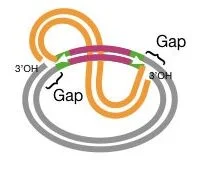
- In this step, strand transfer occurs and a branched DNA molecule is formed. The 3′ ends produced is joined covalently with the 5′ ends of the target site. The 5′ ends of the transposon is still attached to the donor DNA. It is the step that moves the replication strand and creates the intermediate for gap filling.
- The strand transfer complex now produces two replication forks. In this step the 3′ end of the target is cleaved, and it works as a primer. The host replication machinery is formed at the fork junction and replication proceeds from one end to the other end.
- At completion of replication, two DNA molecules is formed containing the transposon. It is the process where short target site duplications is produced which flank the transposon and these sequences move with the element to new positions. Frequent deletions and duplications is common in this mechanism.
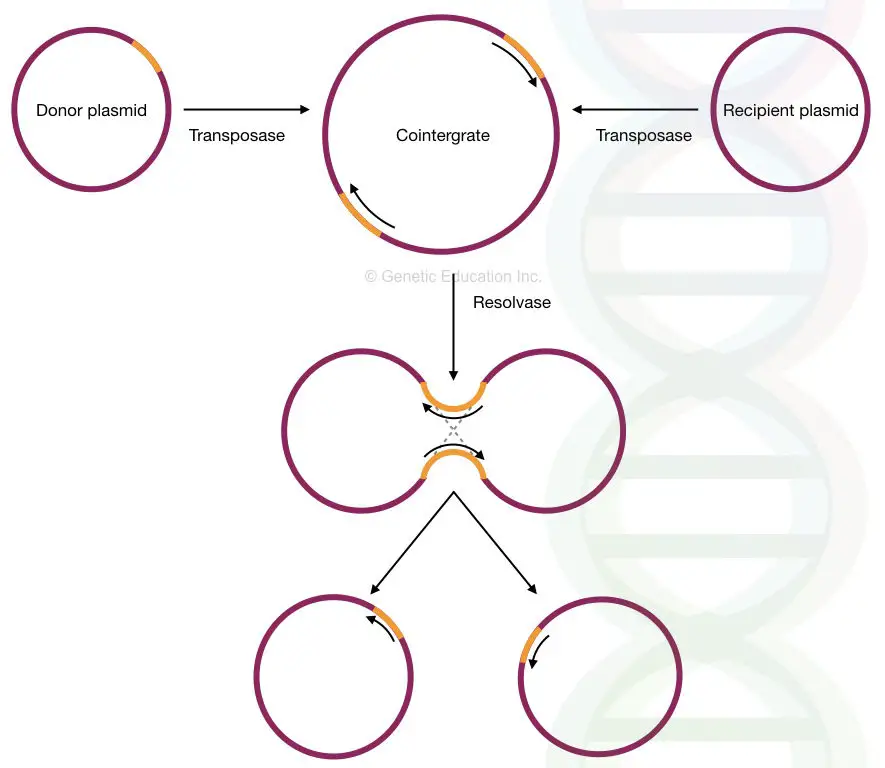
- Replicative transposition occurs through a cointegrate structure in plasmid DNA. It begins when donor and recipient plasmid fuse together. The transposase activity produces a co-integrate. Inside the cointegrate, the transposon replicates and forms a second copy in same orientation.
- Recombination occurs between the two transposon copies in the cointegrate. This step takes place at the res site and it is controlled by transposon and resolvase. Resolvase acts as the bacterial protein which limits transposition and separates the two molecules.
- After resolution, two plasmids is produced each containing a copy of the transposon. It completes the replicative transposition process and both plasmids now carry the duplicated element.
Replicative Transposition of Retrotransposons
- Retrotransposons use a replicative strand transfer mechanism but with an RNA intermediate.
It is the process where reverse transcription replaces direct replication, so the movement occurs through RNA. - In the first stage, RNA is synthesized from the retrotransposon DNA.
The cellular RNA polymerase transcribes the full-length RNA, and the LTR (long terminal repeat) acts as promoter but does not code any protein. - The transcriptional machinery forms the complete RNA molecule from LTR to LTR.
It is the step which prepares the template for reverse transcription. - Reverse transcription now synthesizes DNA from the RNA molecule.
It is the process where cDNA is made, but no copy stays at the original donor site, so only the new cDNA moves. - In the next stage, the cDNA recognises integrase and both ends is assembled with the enzyme.
Integrase binds the cDNA ends and prepares for strand transfer. - Integrase creates a gap at the 3′ ends of the host DNA.
Some nucleotides is removed at these 3′ positions, which is similar to DNA transposon strand transfer. - The integrase enzyme inserts the newly formed DNA into the host genome.
It is the catalytic step where strand transfer occurs and the retrotransposon DNA becomes part of the target DNA. - The DNA repair system fills the remaining gaps.
The recombination is completed when repair enzymes synthesize the missing nucleotides. - Target site duplication is now formed on both ends of the new copy.
It happens because repair activity is same on both sides, so short direct repeats is produced. - This completes the replicative transposition cycle of retrotransposons and DNA transposons.
TY elements of yeast and Copia elements of Drosophila use this mechanism, but Tn3, Tn5 and phage Mu does not follow it.
Replicative Transposition of Tn3
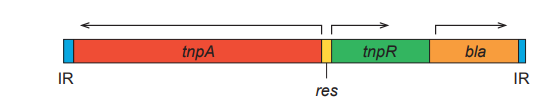
- Tn3 contains the bla gene and two additional genes required for its movement.
It is the transposase gene tnpA and the resolvase gene tnpR. These gene products act in different steps of the process. - Tn3 transposition occurs in two major steps.
Each step is controlled by a different Tn3 gene product, showing that the mechanism is divided into two phases. - The donor plasmid carrying Tn3 interacts with the target DNA.
The target can be another plasmid but sometimes it can also be phage DNA or the bacterial chromosome. - First step is formation of a cointegrate.
It is produced when the donor and target plasmids join together through replication of Tn3. This structure connects both plasmids into one continuous DNA molecule. - Formation of cointegrate requires the tnpA gene product.
The transposase helps the joining reaction and brings together the four DNA strands involved in this process. The replication across Tn3 produces one copy in each plasmid. - The cointegrate is the intermediate of the reaction.
Evidence for this is that tnpR mutants cannot resolve cointegrates, so cointegrates remain as the final product when resolution cannot occur. - Second step is resolution of the cointegrate into two separate molecules.
It is the recombination event between the res sites located within Tn3. This creates two plasmids, each carrying one copy of Tn3. - Resolution is catalyzed by the tnpR gene product (resolvase).
Resolvase acts specifically at the res site and separates the combined DNA into its two components. - If the tnpR gene is defective, cointegration remains unresolved.
It proves that resolvase is required for completion of the transposition cycle. - Resolution can still occur if a functional tnpR is supplied from another DNA molecule.
It shows that recombination at res is dependent on resolvase but does not require it to originate from the same plasmid. - This two-step mechanism defines Tn3 transposition.
It is the process where replicative cointegration is followed by site-specific resolution, producing two molecules each containing Tn3.
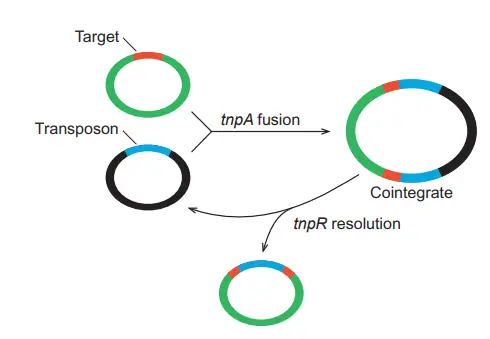
2. Conservative or Nonreplicative transposition (or “cut and paste”)
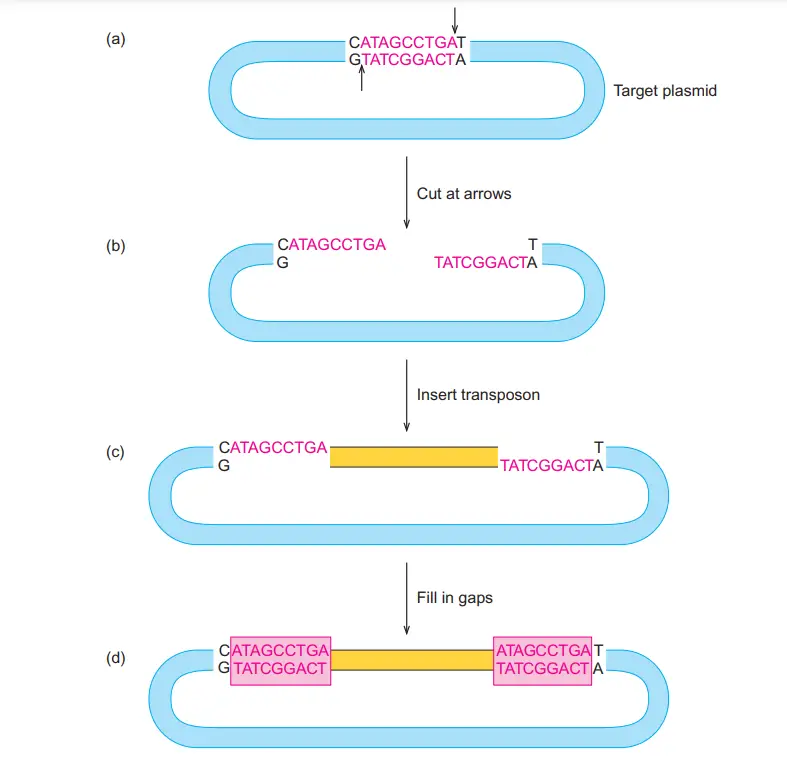
- Some transposons like Tn10 move without forming any new copy. It is the process where the element leaves the donor DNA completely and appears at a new target site.
- The initial step may begin similar to replicative transposition. It is thought that the donor and target DNA strands are cut and joined in the same early manner, forming the primary strand exchange.
- After the first cut-and-join action, a different event now occurs. New nicks is produced on both sides of the transposon within the donor DNA. These nicks free the element from the donor site.
- The transposon remains attached only to the target DNA. It is the point where the donor DNA is left with a gap, while the transposon is covalently linked to the new site.
- The nicks in the target DNA can be sealed by repair enzymes. When this occurs, the transposon becomes fully integrated into the target DNA forming recombinant DNA.
- The donor DNA is left with a break between its two strands. The DNA segment without the transposon may be degraded or sometimes the gap is repaired by the host repair system.
- This completes the nonreplicative or cut-and-paste mechanism. It is the movement where the transposon does not replicate but only changes its position by excision and insertion.
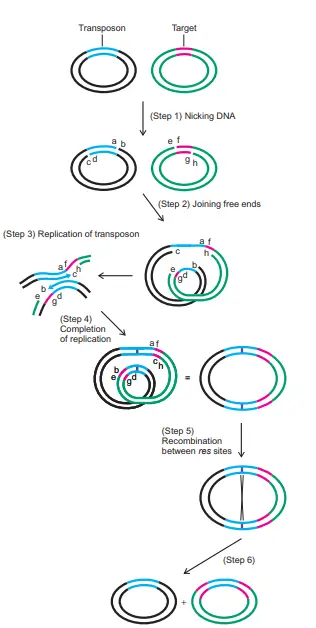
About the above Image
Step 1: involves nicking the two plasmids to create the free ends labelled a-h.
Step 2: Join the ends a and f, as well as g and d. This frees up b, c, e, and h.
Step 3: Two of the remaining free ends (b and c) function as primers for DNA replication, as depicted by the enlargement of the replicating area in Step 3.
Step 4: Continue replicating until the ends of b and c reach e and h, respectively. These ends are ligated in order to finish the cointegrate. Observe that the entire transposon has been duplicated (blue). The paired res sites (purple) are presented for the first time, despite the existence of a single res site in prior rounds. The cointegrate is depicted with a loop so that its derivation from the preceding figure is more apparent; however, if the loop were removed, the cointegrate would resemble the one in Figure 1. (shown here at right).
Step Five and six (resolution): A crossover occurs between the two res sites in the two copies of the transposon, resulting in the formation of two separate plasmids, each of which contains a copy of the transposon.
Mechanism of Nonreplicative transposition
- The mechanism begins with the same two steps seen in replicative transposition.
It is the stage where the donor and target DNA is brought together by the transposase, and the initial cleaving and joining reaction is formed. - The intermediate structure formed is identical to the structure present between step two and three of replicative type.
It shows that both mechanisms share the same early arrangement before the divergence occurs. - Subsequent nicks now appear at specific positions flanking the transposon in the donor DNA.
These nicks release the donor plasmid without the transposon, and only the target DNA remains connected with the element. - The transposon stays covalently attached to the target DNA.
It is the step that fixes the direction of movement because the donor DNA is freed from the complex. - The gaps and nicks in the target DNA are filled and sealed by the repair enzymes.
It completes the integration step and secures the transposon in the target plasmid. - The broken ends of the donor plasmid may or may not rejoin.
In any case, the transposon has been removed from the donor DNA, and the plasmid is left without its original element. - This completes the nonreplicative or cut-and-paste transposition mechanism.
It is the movement where no new copy is generated, and the element is simply excised and inserted at the new site.
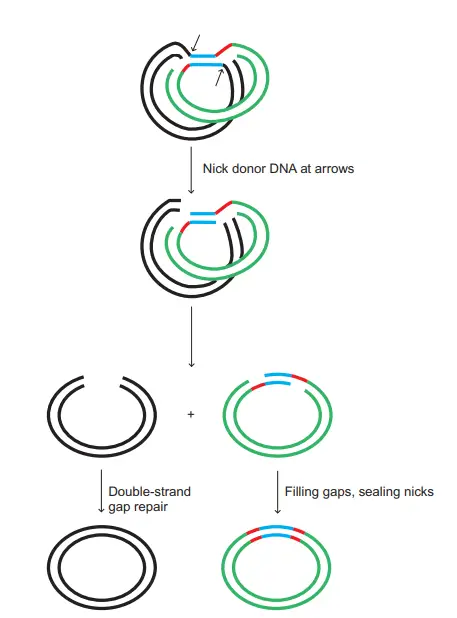
3. Retro-transposons
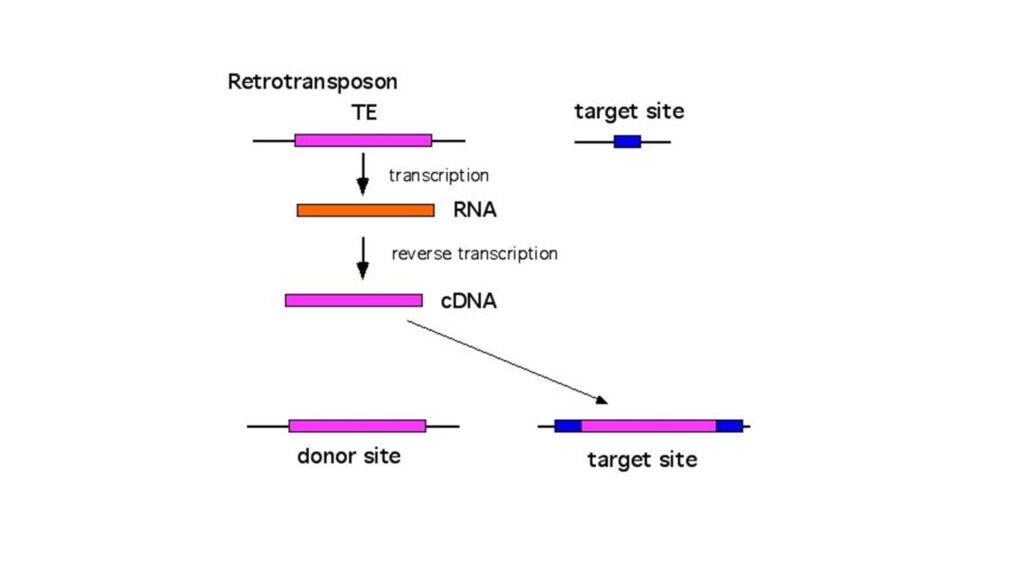
- Retro-transposons are transposons whose RNA is reverse-transcribed into DNA by the reverse transcriptase enzyme, and whose newly synthesised DNA is placed into a new location on a chromosome.
- This type of transposition is referred to as retro-transposition.
- Some of these retro-transposons are connected to retro viruses and utilise their reverse transcriptase enzyme for transposition; these transposable elements are referred to as retroposons.
- Only Eukaryotic organisms possess retro-transposons.
Insertion Sequences: The Simplest Bacterial Transposons
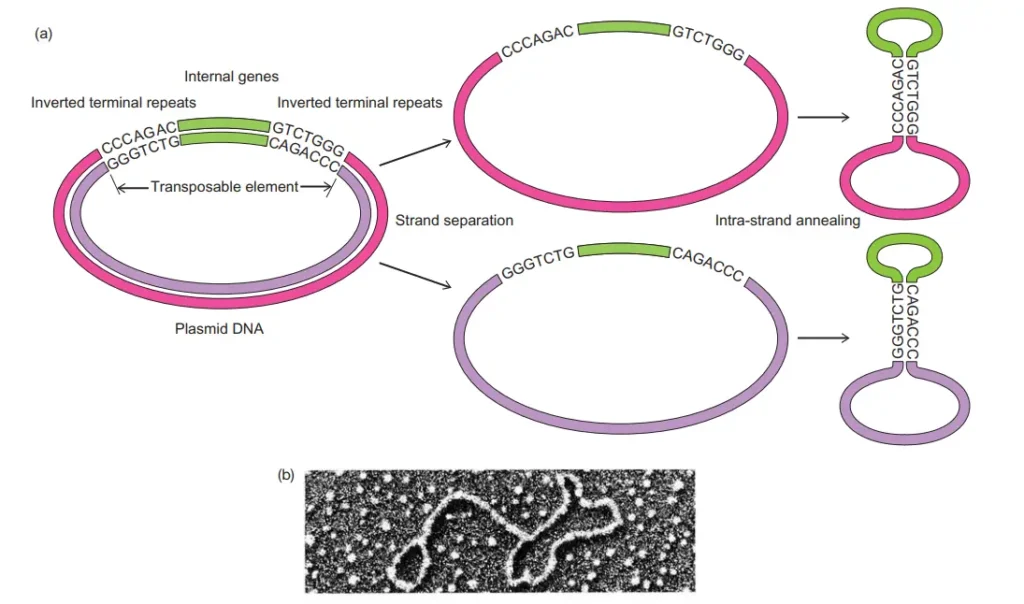
- Insertion sequences contain only the parts required for transposition. It is the simplest type of bacterial transposon and mostly includes the ends and the coding region for the necessary enzymes.
- The ends of an insertion sequence are special inverted repeat sequences. One end is an inverted copy of the other end. For example, if one end is 5′-ACCGTAG, the other will be 3′-CTACGGT. These examples only show the concept.
- Most IS elements have inverted repeats of around 15–25 bp. IS1 has inverted repeats of 23 bp, but in some larger transposons the inverted repeats may be hundreds of base pairs long.
- The internal region of an IS element encodes proteins needed for transposition. It usually encodes at least one transposase. Mutations in this region stop the IS element from moving, showing these proteins are essential.
- Inverted repeats at the ends can be demonstrated experimentally. Cohen separated strands of a recombinant plasmid and predicted that the inverted repeats on one strand would base-pair to form a stem–loop structure. The stem region is double-stranded DNA and the loop is single-stranded.
- The stem–loop structure is visible under the electron microscope. It confirms that inverted repeats exist at the ends of IS elements.
- Insertion sequences also produce short direct repeats in the target DNA. These repeats are not present earlier. They appear only after insertion and show that the transposase does not cut the target DNA at adjacent positions.
- The distance between the staggered cuts determines the length of the direct repeats. It depends on how the IS element cuts the DNA. For example, IS1 transposase cuts 9 bp apart, so the direct repeats are also 9 bp long.
- These direct repeats flank the transposon in the final inserted position. It is one important feature shared by insertion sequences as well as more complex transposons.
- Mechanisms of DNA Transposition. (n.d.). Mobile DNA III, 531–553. doi:10.1128/microbiolspec.mdna3-0034-2014
- Bourque, G., Burns, K.H., Gehring, M. et al. Ten things you should know about transposable elements. Genome Biol 19, 199 (2018). https://doi.org/10.1186/s13059-018-1577-z
- Muñoz-López M, García-Pérez JL. DNA transposons: nature and applications in genomics. Curr Genomics. 2010 Apr;11(2):115-28. doi: 10.2174/138920210790886871. PMID: 20885819; PMCID: PMC2874221.
- https://www.onlinebiologynotes.com/transposable-elements-characteristics-and-mechanisms-of-transposition/
- https://www.edx.org/course/molecular-biology-part-2-transcription-and-transposition
- http://what-when-how.com/molecular-biology/transposition-molecular-biology/
- https://pdb101.rcsb.org/motm/84
- https://www.annualreviews.org/doi/abs/10.1146/annurev.cb.06.110190.001501?journalCode=cellbio.1
- https://www.slideshare.net/vivekaiden/transposones
- https://www.ibiology.org/genetics-and-gene-regulation/transposable-elements/
- http://www.bio.brandeis.edu/classes/biol122a/Transposition.htm
- https://www.notesonzoology.com/plants/transposable-elements-in-different-plants-biology/5342
- https://www.classcentral.com/course/edx-molecular-biology-part-2-transcription-and-transposition-4072
- https://journals.asm.org/doi/full/10.1128/microbiolspec.MDNA3-0034-2014
- https://www.biologyonline.com/dictionary/transposition
9. Transposition of DNA. (2021, July 20). The Pennsylvania State University. - https://bio.libretexts.org/@go/page/364
- http://www.sci.sdsu.edu/~smaloy/MicrobialGenetics/problems/transposons/transposition/
- https://en.wikipedia.org/wiki/Transposable_element
- https://www.biologydiscussion.com/genetics/transposition-meaning-and-mechanism-genetics/65170
- https://www.nature.com/scitable/topicpage/transposons-the-jumping-genes-518/
- https://www.sciencedirect.com/topics/biochemistry-genetics-and-molecular-biology/dna-transposition
- https://geneticeducation.co.in/replicative-transposition-of-dna-transposons-and-retrotransposons/
- https://en.wikipedia.org/wiki/Transposition
- https://viralzone.expasy.org/4017