Gram-negative bacteria are a diverse group of microorganisms known for their unique cell structure, which plays a key role in how they interact with the environment and cause infections. Unlike Gram-positive bacteria, Gram-negative bacteria have a thin peptidoglycan layer sandwiched between two membranes. The outer membrane contains lipopolysaccharides (LPS), which can trigger strong immune responses in humans, sometimes leading to severe symptoms like fever or shock. This double-layer structure also makes Gram-negative bacteria more resistant to antibiotics, contributing to their reputation as challenging pathogens in healthcare settings.
Common examples of Gram-negative bacteria include E. coli, Salmonella, and Pseudomonas aeruginosa, which are linked to illnesses like urinary tract infections, food poisoning, and pneumonia. Their antibiotic resistance is partly due to the outer membrane acting as a barrier, limiting drug penetration, and specialized pumps that eject harmful substances. Understanding Gram-negative bacteria is critical for developing effective treatments and preventing infections, especially in hospitals where they often thrive. Researchers continue to study their adaptability and mechanisms to combat the growing threat of drug-resistant strains. By prioritizing hygiene, proper antibiotic use, and innovative therapies, we can better manage risks associated with these resilient microbes.
What are Gram-negative bacteria?
- Gram-negative bacteria are a category of prokaryotic microorganisms distinguished by their failure to maintain the crystal violet stain during Gram staining, leading to a pink or red coloration from the absorption of the counterstain, safranin.
- They have a distinctive cell wall architecture consisting of a slender peptidoglycan layer located between an inner cytoplasmic membrane and an outside membrane that contains lipopolysaccharides (LPS).
- The outer membrane functions as a protective barrier, enhancing the bacteria’s resistance to certain medicines and detergents.
- The lipopolysaccharide component, especially lipid A, functions as an endotoxin, eliciting robust immune responses and potentially resulting in septic shock following bacterial lysis.
- Gram-negative bacteria are ubiquitous in many habitats and encompass both non-pathogenic and pathogenic species. Prominent pathogenic genera encompass Escherichia, Salmonella, Neisseria, and Pseudomonas.
- These bacteria are implicated in several illnesses, including pneumonia, bloodstream infections, urinary tract infections, and meningitis, especially within hospital environments.
- The structural characteristics of Gram-negative bacteria, particularly the outer membrane, enhance their inherent resistance to several antibiotics, presenting considerable obstacles in therapeutic management.
- The Gram staining technique, established by Hans Christian Gram in 1884, is a crucial tool for bacterial classification and diagnosis, differentiating bacteria based on cell wall composition.
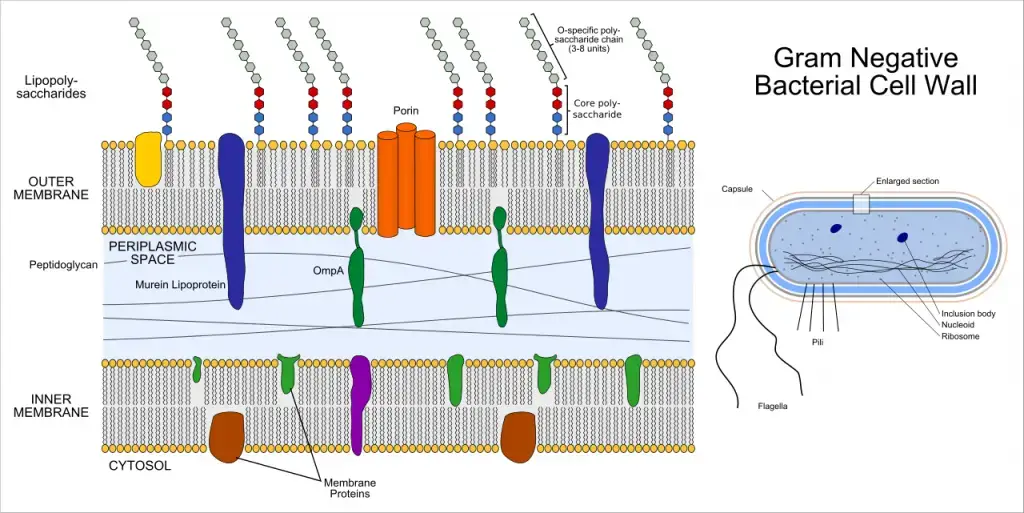
Characteristics gram-negative bacteria
- After counterstaining with safranin, gram-negative bacteria show pink or red instead of retaining the crystal violet dye in Gram staining.
- Their cell envelope has an inner cytoplasmic membrane, a thin peptidoglycan layer in the periplasmic space, and an outside membrane heavy in lipopolysaccharides.
- Hydrolytic enzymes and binding proteins fundamental for food uptake and cell wall building abound in the periplasmic region.
- Three sections make up lipopolysaccharides found in the outer membrane.
- fat An attaches LPS to the membrane and functions as an endotoxin able to cause septic shock.
- Main oligosaccharide links lipid A to the O antigen.
- O antigen polysaccharide helps immune escape and differs depending on species.
- Water-filled channels formed by outer membrane porins control small hydrophilic molecule inflow and help to explain antibiotic resistance.
- Thin (probably 7–10 nm), the peptidoglycan layer gives structural support but less stiffness than in Gram-positive bacteria.
- Many antibiotics and detergues are blocked by the outer membrane barrier, therefore giving natural resistance.
- Many species have surface features like pili and flagella that help adhesion, movement, and biofilm development.
- Many times carrying plasmids encoding antibiotic resistance genes and virulence factors, genes help to promote horizontal gene transfer and quick adaption.
- Playing important roles in virulence, specialized secretion systems (type I–VI) cross the cell membrane to export proteins and poisons straight into host cells or the environment.
- Aerobic and anaerobic respiration as well as fermentation help to colonize several ecological niches by means of metabolic capacities.
- Escherichia, Salmonella, Pseudomonas, Neisseria, and Klebsiella are among common pathogenic genera that cause illnesses including sepsis, urinary tract infections, and pneumonia.
- From rods and cocci to vibrios and spirilla, usually 0.2–1µm in width, cell forms range from
- In clinical and scientific microbiology, Hans Christian Gram’s Gram staining method created in 1884 is still basic for bacterial taxonomy.
- Common in soil, water, and animal gastrointestinal systems, Gram-negative bacteria may operate as opportunistic or main pathogens and help to cycle nutrients.
Gram-negative bacteria cell wall Structure
- Three separate layers make up the gram-negative bacterial cell envelope: the inner cytoplasmic membrane, a thin peptidoglycan layer in the periplasmic region, and an outside membrane with lipopolysaccharide.
- Comprising phospholipid bilayers with proteins for energy production, transport, and signaling, the inner cytoplasmic membrane
- Greater than Gram positive bacteria, the periplasmic space between the inner and outer membranes is a gel-like compartment filled in enzymes for nutrition processing and cell wall construction.
- Thin 2–7 nm single mesh like polymer of N acetylglucosamine and N acetylmuramic acid provides structural support from the peptidoglycan layer.
- About 7–8 nm thick, the outer membrane is an asymmetric bilayer with phospholipids in the inner leaflet and lipopolysaccharide in the outside leaflet.
- Three sections define lipopolysaccharide.
- fat As endotoxin, anchors LPS to the membrane.
- Core oligosaccharides bind lipid A to the O antigen.
- O antigen polysaccharide differs among species to help immune escape.
- Braun’s lipoprotein preserves envelope integrity and bacterial form by tethering the outer membrane to the peptidoglycan layer.
- Beta barrel proteins called outer membrane porins provide water-filled channels for passive diffusion of hydrophilic molecules up to roughly 600 daltons.
- Porins and other outer membrane proteins act as receptors for bacteriophages and bacteriocins.
- Key functions of bacterial pathogenicity include secretion systems spanning the envelope export proteins and toxins.
- The cell envelope barrier controls the entrance of antibiotics and detergues therefore facilitating natural resistance mechanisms.
- Variations in LPS O antigen and outer membrane proteins help to enable antigenic diversity and host immune response evasion.
- Gram negative bacteria can resist strong turgor pressure and external challenges by means of envelopes flexibility and strength.
- Assembly of outer membrane proteins necessary for envelope function by autotransporters and lipoprotein export machinery
The Periplasmic Space in Gram-Negative Bacteria
The periplasmic space in Gram-negative bacteria is a critical and complex component, situated between the outer membrane and the cytoplasmic membrane. This space plays a pivotal role in the bacteria’s functionality and survival.
- Structure and Composition:
- Location: The periplasmic space is found between the outer membrane and the cytoplasmic membrane. It is more prominent in Gram-negative bacteria than in Gram-positive bacteria.
- Content: This space contains a gel-like substance known as periplasm, which is rich in proteins, enzymes, and other essential molecules.
- Proteins and Enzymes:
- Hydrolytic Enzymes: These enzymes break down nucleic acids and phosphorylated molecules, aiding in the degradation of complex nutrients into simpler, absorbable forms.
- Binding Proteins: These proteins play an active role in the transport of materials into the bacterial cell, ensuring efficient nutrient acquisition.
- Peptidoglycan Synthesis:
- Enzymatic Activity: Enzymes within the periplasmic space are involved in the synthesis and remodeling of peptidoglycan, which is crucial for maintaining cell wall integrity and shape.
- Toxin Modification:
- Detoxifying Enzymes: The periplasmic space contains enzymes that modify and neutralize toxic elements, protecting the bacterial cell from harmful substances.
- Functional Roles:
- Nutrient Acquisition: The presence of hydrolytic enzymes and binding proteins ensures that the bacteria efficiently absorb necessary nutrients from their environment.
- Cell Wall Maintenance: Enzymes involved in peptidoglycan synthesis maintain the structural integrity and shape of the bacterial cell wall.
- Protection: The detoxifying enzymes present in the periplasmic space provide a defense mechanism against toxic elements that could otherwise damage the cell.
- Importance in Bacterial Physiology:
- The periplasmic space is essential for the bacteria’s survival and adaptation in various environments. It supports crucial processes such as nutrient uptake, cell wall synthesis, and protection against environmental stressors.
Peptidoglycan in Gram-Negative Bacteria
- In Gram-negative bacteria, peptidoglycan is a thin mesh of alternating N‑acetylglucosamine and N‑acetylmuramic acid strands cross‑linked by short peptides to surround the cytoplasmic membrane.
- By opposing internal turgor pressure, this mesh imparts structure and stiffness and prevents osmotic lysis in hypertonic conditions.
- Biography Liberties Texts
- Early biochemical studies by Weidel and Pelzer used the name murein in 1964; later, Ghuysen used the term peptidoglycan to characterise this macromolecule in 1966.
- Typical L‑alanine, D‑glutamate, meso‑diaminopimelic acid, and D‑alanine in Escherichia coli each glycan strand consists of β(1→4) connected N‑acetylglucosamine and N‑acetylmuramic acid residues with muramic acid bearing a peptide stem.
- Usually two to seven nanometers thick, the sacculus in Gram-negative bacteria is a single thin layer with less cross-link density than in Gram-positive bacteria, therefore offering both strength and flexibility.
- In Gram-negative bacteria, peptidoglycan finds its place in the periplasmic gap between the inner cytoplasmic membrane and the outside lipopolysaccharide-rich membrane.
- Gram-negative bacteria have a thin peptidoglycan layer and an extra outer bilayer that acts as a permeability barrier unlike Gram-positive bacteria with thick multilayered peptidoglycan and without an outside membrane.
- Two steps of biosynthesis follow cytoplasmic assembly of UDP‑NAG and UDP‑NAM pentapeptide precursors under Glm and Mur enzymes followed by membrane connection of lipid II, translocation by flippase, transglycosylase polymerization, and DD‑transpeptidase cross‑linking.
- Beta-lactam antibiotics target the thin peptidoglycan sacculus primarily; changes in its production and structure are fundamental to antibiotic resistance mechanisms in Gram-negative bacteria.
- Advances in peptidoglycan research include fabrication of muropeptide probes, investigation of cell wall remodeling enzymes, and development of new antibacterial methods targeting special Gram-negative production routes.
The Outer Membrane and Lipopolysaccharides in Gram-Negative Bacteria
The outer membrane of Gram-negative bacteria is a complex and vital structure. Situated above the thin peptidoglycan layer, it serves multiple critical functions in protecting and maintaining the bacterial cell.
- Structure of the Outer Membrane:
- Composition: The outer membrane is primarily composed of lipopolysaccharides (LPS), phospholipids, lipoproteins, and surface proteins.
- Braun’s Lipoprotein: This protein covalently binds to the peptidoglycan and anchors itself to the outer membrane via its hydrophobic ends. It provides a strong linkage between the peptidoglycan and the outer membrane.
- Functional Components:
- Adhesion Sites: These sites on the outer membrane allow cell contact and membrane fusion, facilitating substance entry into the cell.
- Porin Proteins: Porins are integral proteins that form channels allowing the passive diffusion of small molecules like glucose across the outer membrane. Larger molecules, such as Vitamin B12, require specific carriers.
- Lipopolysaccharides (LPS):
- Structure: LPS are large, complex molecules comprising three main parts: Lipid A, core polysaccharides, and the O side chain (O antigen).
- Lipid A: This component consists of two glucosamine sugar derivatives, each with three fatty acids and pyrophosphate. It embeds in the membrane and contributes to the endotoxic properties of LPS.
- Core Polysaccharides: These connect Lipid A to the O side chain and help stabilize the LPS structure.
- O Side Chain (O Antigen): Extending outward from the core, the O side chain consists of sugars that vary among bacterial strains, aiding in immune evasion by altering surface antigens.
- Functions of Lipopolysaccharides:
- Barrier Function: LPS protect the cell wall from external threats, including antibiotics and bile salts, by forming a barrier.
- Structural Stability: The negative charge of LPS contributes to the overall negative charge of the cell surface, stabilizing the membrane structure.
- Endotoxic Properties: Lipid A is toxic to host organisms, acting as an endotoxin that can trigger strong immune responses.
- Role in Antibiotic Resistance:
- The outer membrane’s structure significantly impedes the entry of harmful substances. This barrier function is crucial in preventing antibiotics and other toxic agents from penetrating and disrupting the cell.
- Preventing Component Loss:
- Besides protecting against external threats, the outer membrane also prevents the loss of vital components from the periplasmic space, ensuring the cell retains essential enzymes and nutrients.
Lipoprotein layer
The layer of lipoprotein is comprised of Braun’s lipoprotein. Braun’s lipoprotein can be described as a small lipoprotein which is covalently attached to the peptidoglycan underneath and is embedded within the membrane’s outer part through its hydrophobic ends. lipoprotein helps to stabilize the outer layer of the cell wall that is Gram-negative.
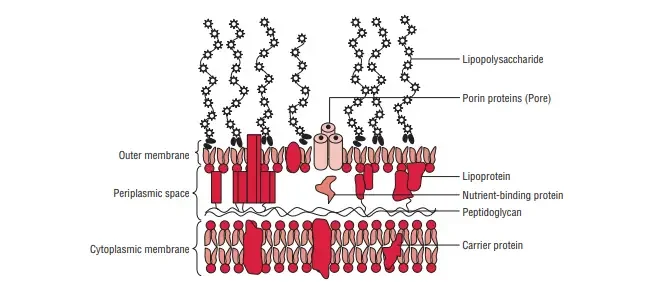
The Outer Membrane in Gram-Negative Bacteria
The outer membrane of Gram-negative bacteria is a critical structure with a unique two-layered design. This membrane plays a vital role in maintaining the integrity and functionality of the bacterial cell.
- Structure of the Outer Membrane:
- Bilayer Design: The inner layer of the outer membrane is similar to the cell membrane, composed primarily of phospholipids. However, the outer layer contains a unique component called lipopolysaccharide (LPS).
- Adhesion: The outer membrane and the plasma membrane are connected at multiple points within the Gram-negative cell wall.
- Key Proteins in the Outer Membrane:
- Porins: These are protein molecules that form special channels in the outer membrane. They serve several functions:
- Diffusion Channels: Porins allow the diffusion of low-molecular-weight hydrophilic substances such as amino acids, sugars, and certain ions.
- Selective Barrier: They exclude hydrophobic molecules, thereby protecting the cell.
- Defense Mechanism: Porins contribute to the cell’s defense against harmful substances.
- Outer Membrane Proteins (OMPs): These include various proteins that facilitate different functions:
- OmpC, OmpD, OmpF, PhoE, LamB: These major proteins are involved in the transmembrane transport of maltose and maltodextrins.
- Tsx: Acts as the receptor for T6 bacteriophage and is responsible for the transmembrane transport of nucleosides and certain amino acids.
- OmpA: This protein binds the outer membrane to the peptidoglycan layer and acts as the receptor for the sex pilus during F-mediated bacterial conjugation.
- Porins: These are protein molecules that form special channels in the outer membrane. They serve several functions:
- Specialized Transport Functions:
- The outer membrane contains proteins that transport specific molecules, such as Vitamin B12 and iron-siderophore complexes.
- Additionally, it houses minor proteins like phospholipases, enzymes, and proteases.
- Functional Significance:
- Barrier and Protection: The outer membrane acts as a barrier, preventing the entry of harmful substances, including antibiotics and bile salts.
- Structural Stability: The unique composition of the outer membrane, including LPS, contributes to the overall stability of the bacterial cell wall.
- Nutrient Transport: Porins and other OMPs facilitate the transport of essential nutrients into the cell, ensuring proper cellular function.
Structure of a lipopolysaccharide
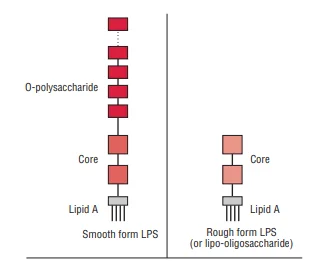
Lipopolysaccharides (LPS) are complex molecules found in the outer membrane of Gram-negative bacteria. These molecules are critical for the bacterial cell’s integrity and interaction with its environment. The structure of LPS consists of three main components: lipid A, the core oligosaccharide, and the O polysaccharide, also known as the O-antigen.
- Lipid A:
- Composition: Lipid A is composed of a phosphorylated disaccharide unit. Attached to this unit are several long-chain fatty acids. A notable feature is the presence of hydroxymyristic acid, a fatty acid associated with the endotoxic properties of LPS.
- Variability: While the structure of lipid A can vary among different Gram-negative species, it remains relatively consistent within the same species. This consistency is crucial for the stability and function of the outer membrane.
- Core Oligosaccharide:
- Structure: The core oligosaccharide includes distinctive sugars such as ketodeoxyoctanoic acid (KDO) and heptose, which link to lipid A. This component is genus-specific, common to all Gram-negative bacteria.
- Lipooligosaccharides (LOS): In some bacteria, such as Neisseria meningitidis, Neisseria gonorrhoeae, Haemophilus influenzae, and Haemophilus ducreyi, the core oligosaccharide forms smaller glycolipids known as LOS. These possess shorter, branched glycans.
- Antigenic Diversity: LOS exhibit significant antigenic and structural diversity, even within a single strain. The N-acetyllactosamine residue on LOS is immunochemically similar to human erythrocyte I antigen. Sialylation of this residue offers the bacteria molecular mimicry advantages, helping them evade the host’s immune response.
- O Polysaccharide (O-Antigen):
- Extension and Composition: Extending from the core oligosaccharide, the O polysaccharide contains various unique sugars. Its composition varies between bacterial strains, giving each strain specific antigenic properties.
- Immune Evasion: The O-antigen is exposed to the host’s immune system. Gram-negative bacteria can alter their O side chains to evade detection by the host’s defenses. This adaptability allows the bacteria to persist and thrive in hostile environments.
Gram-Negative Bacteria: Pathologies and Clinical Significance
Gram-negative bacteria are part of the normal human flora, but some can cause severe infections. These infections range from community-acquired to nosocomial (hospital-acquired) infections. If not detected and treated promptly, they can lead to serious health issues, even death. Below is a list of some Gram-negative bacteria and the clinical features associated with the diseases they cause.
1. Neisseria gonorrhoeae
- Genitourinary Tract Infections: In males, this bacterium infects the urethra, leading to purulent urethral discharge and painful urination. In females, it affects the vagina and endocervix, possibly progressing to the uterus, causing salpingitis, pelvic inflammatory disease (PID), and fibrosis. Salpingitis can lead to infertility.
- Renal Infection: Men may experience constipation, painful defecation, and purulent discharge.
- Pharyngitis: Purulent pharyngeal exudation can cause pharyngitis.
- Ophthalmia Neonatorum: Newborns can acquire this infection during birth, leading to eye infections.
- Disseminated Infection: Symptoms include fever, painful purulent arthritis, and small scattered pustules on the skin with an erythematous base. Necrosis may develop.
2. Neisseria meningitidis
- Meningitis: This bacterium can cause meningococcemia if it invades the bloodstream, associated with high fever. It can also spread to the brain, causing purulent meningitis with fever, severe headaches, joint aches, and a petechial or purpuric rash.
- Septicemia: Rapid onset of septicemia can progress to fulminant septicemia and shock, especially in children (Waterhouse-Friderichsen syndrome).
3. Escherichia coli
- Intestinal Diseases: These include enterotoxigenic (ETEC), enteropathogenic (EPEC), enterohemorrhagic (EHEC), enteroinvasive (EIEC), and enteroaggregative (EAEC) strains, all associated with diarrhea (watery or bloody).
- Extraintestinal Diseases: E. coli can cause urinary tract infections (UTIs), neonatal meningitis, and nosocomial infections like sepsis, bacteremia, endotoxic shock, and pneumonia.
4. Salmonella spp.
- Enteric and Typhoid Fever: Characterized by fever, abdominal pain, chills, sweats, headache, anorexia, weakness, sore throat, cough, myalgia, and either diarrhea or constipation.
- Gastroenteritis (Salmonellosis): Symptoms include nausea, vomiting, and non-bloody diarrhea.
- Bacteremia: Associated with abdominal infections, osteomyelitis, and septic arthritis.
5. Campylobacter jejuni
- Intestinal and Extraintestinal Disease: Symptoms include fever, headache, myalgia, abdominal cramping, and diarrhea, which may be bloody. It commonly causes traveler’s diarrhea and pseudoappendicitis.
- Bacteremia: Often transient, it occurs most frequently in infants and older adults.
6. Shigella dysenteriae
- Shigellosis (Bacillary Dysentery): Characterized by diarrhea with blood, mucus in stool, and painful abdominal cramping.
7. Vibrio cholerae
- Cholera: Associated with profuse watery diarrhea, leading to massive fluid and electrolyte loss.
8. Helicobacter pylori
- Acute Gastritis: Causes diarrhea and epigastric discomfort.
- Ulcers: Can cause both duodenal and gastric ulcers. Persistent ulceration may lead to mucosa-associated lymphoid tumors.
9. Klebsiella pneumoniae
- UTI and Nosocomial Infections: Can cause urinary tract infections and bacteremia in hospital settings.
10. Pseudomonas aeruginosa
- Opportunistic Infections: Common in wounded patients and those with catheters or respirators.
- Eye Infections: Keratitis and endophthalmitis following injuries.
- Skin and Respiratory Infections: Causes skin wound infections and pneumonia symptoms.
- Gastrointestinal Infections: Associated with diarrhea and necrotic enterocolitis in infants.
- Systemic Infections: Includes septicemia, pneumonia, bone and joint infections, CNS infections, and soft tissue infections in hospitalized patients.
| Gram-Negative Bacteria | Pathologies | Clinical Features |
|---|---|---|
| Neisseria gonorrhoeae | Genitourinary Tract Infections | Purulent urethral discharge, painful urination in males; vagina and endocervix infection in females, progressing to salpingitis, PID, and fibrosis; infertility; renal infection in men with constipation, painful defecation, and purulent discharge; pharyngitis; ophthalmia neonatorum in newborns; disseminated infection with fever, purulent arthritis, pustules with erythematous base, and possible necrosis. |
| Neisseria meningitidis | Meningitis and Septicemia | Rapid onset of meningococcemia with high fever; purulent meningitis with severe headaches, joint aches, petechial or purpuric rash; septicemia progressing to fulminant septicemia and shock, especially in children (Waterhouse-Friderichsen syndrome). |
| Escherichia coli | Intestinal Diseases and Extraintestinal Diseases | Diarrhea (watery or bloody) from ETEC, EPEC, EHEC, EIEC, EAEC; UTIs, neonatal meningitis, sepsis, bacteremia, endotoxic shock, pneumonia. |
| Salmonella spp. | Enteric and Typhoid Fever, Gastroenteritis, Bacteremia | Fever, abdominal pain, chills, sweats, headache, anorexia, weakness, sore throat, cough, myalgia, diarrhea or constipation; nausea, vomiting, non-bloody diarrhea; abdominal infections, osteomyelitis, septic arthritis. |
| Campylobacter jejuni | Intestinal and Extraintestinal Disease, Bacteremia | Fever, headache, myalgia, abdominal cramping, diarrhea (may be bloody); traveler’s diarrhea, pseudoappendicitis; transient bacteremia, especially in infants and older adults. |
| Shigella dysenteriae | Shigellosis (Bacillary Dysentery) | Diarrhea with blood, mucus in stool, and painful abdominal cramping. |
| Vibrio cholerae | Cholera | Profuse watery diarrhea leading to massive fluid and electrolyte loss. |
| Helicobacter pylori | Acute Gastritis and Ulcers | Diarrhea, epigastric discomfort, duodenal and gastric ulcers; persistent ulceration may lead to mucosa-associated lymphoid tumors. |
| Klebsiella pneumoniae | UTI and Nosocomial Infections | Urinary tract infections, nosocomial bacteremia. |
| Pseudomonas aeruginosa | Opportunistic Infections, Eye Infections, Skin and Respiratory Infections, Gastrointestinal Infections, Systemic Infections | Wound infections from surgeries, eye infections post-injury, skin wound infections, pneumonia, gastrointestinal infections with diarrhea, necrotic enterocolitis in infants, systemic infections including septicemia, pneumonia, bone and joint infections, CNS infections, and soft tissue infections in hospitalized patients. |
Gram-Negative Bacteria and Antimicrobial Agents
| Antibiotic | Mode of Action | Bacterial Agent |
|---|---|---|
| Cephalosporin: Ceftriaxone | Disrupts cell wall synthesis by binding to penicillin-binding proteins and enzymes responsible for peptidoglycan synthesis | Neisseria gonorrhoeae, Neisseria meningitidis, Pseudomonas aeruginosa |
| Tetracycline: Doxycycline | Inhibits protein synthesis by preventing elongation of polypeptides at 30S ribosomes | Neisseria gonorrhoeae |
| Streptogramins | Inhibit protein synthesis by preventing polypeptide elongation at 50S ribosomes | Neisseria gonorrhoeae |
| β-lactam: Penicillin G | Disrupts cell wall synthesis by inhibiting penicillin-binding proteins and enzymes used for peptidoglycan synthesis | Neisseria meningitidis, Pseudomonas aeruginosa |
| Rifampin: Rifamycin | Inhibits nucleic acid synthesis by preventing transcription through binding to DNA-dependent RNA polymerase | Neisseria meningitidis, Escherichia coli |
| Macrolides: Erythromycin, Azithromycin, Clarithromycin | Inhibit bacterial protein synthesis by preventing polypeptide elongation at 50S ribosomes | Neisseria gonorrhoeae, Campylobacter jejuni, Shigella dysenteriae, Helicobacter pylori, Pseudomonas aeruginosa |
| Quinolones: Fluoroquinolones, Ciprofloxacin | Inhibit nucleic acid synthesis by binding to the alpha-subunit of DNA gyrase | Escherichia coli, Salmonella typhi/paratyphi, Campylobacter jejuni, Shigella dysenteriae, Pseudomonas aeruginosa |
| Aminoglycosides | Inhibit protein synthesis by causing premature production of aberrant peptide chains at 30S ribosomes | Escherichia coli (localized and systemic infections) |
| Sulfonamides: Sulfamethoxazole | Act as antimetabolites by inhibiting dihydropteroate synthase, disrupting folic acid synthesis | Escherichia coli (UTIs and systemic diseases) |
| Trimethoprim | Acts as an antimetabolite by inhibiting dihydrofolate reductase, disrupting folic acid synthesis | Escherichia coli (UTIs) |
- Silhavy, T. J., Kahne, D., & Walker, S. (2010). The bacterial cell envelope. Cold Spring Harbor Perspectives in Biology, 2(5), a000414.
- Needham, B. D., & Trent, M. S. (2013). Fortifying the barrier: the impact of lipid A remodelling on bacterial pathogenesis. Nature Reviews Microbiology, 11(7), 467-481.
- Tacconelli, E., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. The Lancet Infectious Diseases, 18(3), 318-327.
- Zgurskaya, H. I., López, C. A., & Gnanakaran, S. (2015). Permeability barrier of Gram-negative cell envelopes and approaches to bypass it. ACS Infectious Diseases, 1(11), 512-522.
- Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., & Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nature Reviews Microbiology, 13(1), 42-51.
- Neidhardt, F. C., Ingraham, J. L., & Schaechter, M. (1990). Physiology of the Bacterial Cell: A Molecular Approach. Sinauer Associates.
- Beveridge, T. J., & Graham, L. L. (1991). Surface Layers of Bacteria. Microbiological Reviews, 55(4), 684-705.
- Rietschel, E. T., & Brade, H. (1992). Bacterial Endotoxins. Scientific American, 267(2), 54-61.
- Salton, M. R. J., & Kim, K. S. (1996). Structure. In: Baron S, editor. Medical Microbiology. 4th edition.
- Galveston (TX): University of Texas Medical Branch at Galveston.
- Willey, J. M., Sherwood, L. M., & Woolverton, C. J. (2020). Prescott’s Microbiology (11th ed.). McGraw-Hill Education.