What is Sequence Alignment?
Sequence alignment is a computational technique used to compare and analyze the similarities and differences between two or more sequences of biological data, such as DNA, RNA, or protein sequences. By aligning sequences, researchers can identify conserved regions, detect mutations, infer evolutionary relationships, and predict functional elements. It involves arranging the sequences in a way that maximizes matches or minimizes mismatches and indels (insertions and deletions). Pairwise alignment focuses on comparing two sequences, while multiple sequence alignment extends this to incorporate three or more sequences. Sequence alignment algorithms assign scores or penalties to determine the most probable evolutionary relationship or functional similarity between the sequences. This technique plays a vital role in fields such as genomics, proteomics, evolutionary biology, drug discovery, and forensic analysis, enabling insights into the structure, function, and evolution of biological sequences.
Two commonly used sequence alignment algorithms are global alignment and local alignment.
Global alignment and local alignment are two fundamental concepts in sequence alignment that serve different purposes and address different alignment scenarios:
- Global Alignment: Global alignment is a type of sequence alignment that aims to align the entire length of two or more sequences. It finds the best alignment between the sequences by considering the entire length of the sequences, from the beginning to the end. Global alignment is suitable when the sequences being compared are expected to have significant similarity throughout their entire lengths.
The Needleman-Wunsch algorithm is commonly used for global alignment. It assigns scores to different alignment possibilities and finds the alignment with the highest score. Global alignment is useful for comparing sequences that share a common evolutionary history, identifying conserved regions, and studying overall sequence similarity.
- Local Alignment: Local alignment, on the other hand, focuses on identifying and aligning regions of similarity or “high-scoring segments” between sequences. It aims to find the best alignment within a specific region of the sequences, disregarding the remaining parts that may not be similar. Local alignment is suitable when the sequences being compared are expected to have significant similarity only in certain regions.
The Smith-Waterman algorithm is commonly used for local alignment. It allows for the identification of local similarities by considering negative scores as zero, thereby finding the alignment with the highest local similarity score. Local alignment is useful for identifying functional domains, detecting conserved motifs, and identifying regions of significance within larger sequences.
In summary, global alignment aligns the entire length of sequences and is suitable for sequences with significant similarity throughout their lengths, while local alignment focuses on aligning specific regions of similarity within sequences and is suitable for sequences with localized similarity. The choice between global and local alignment depends on the specific research question, the expected characteristics of the sequences, and the desired insights from the alignment analysis.
Types of Sequence Alignment
Pairwise alignment and multiple sequence alignment (MSA) are the two primary categories of sequence alignment.
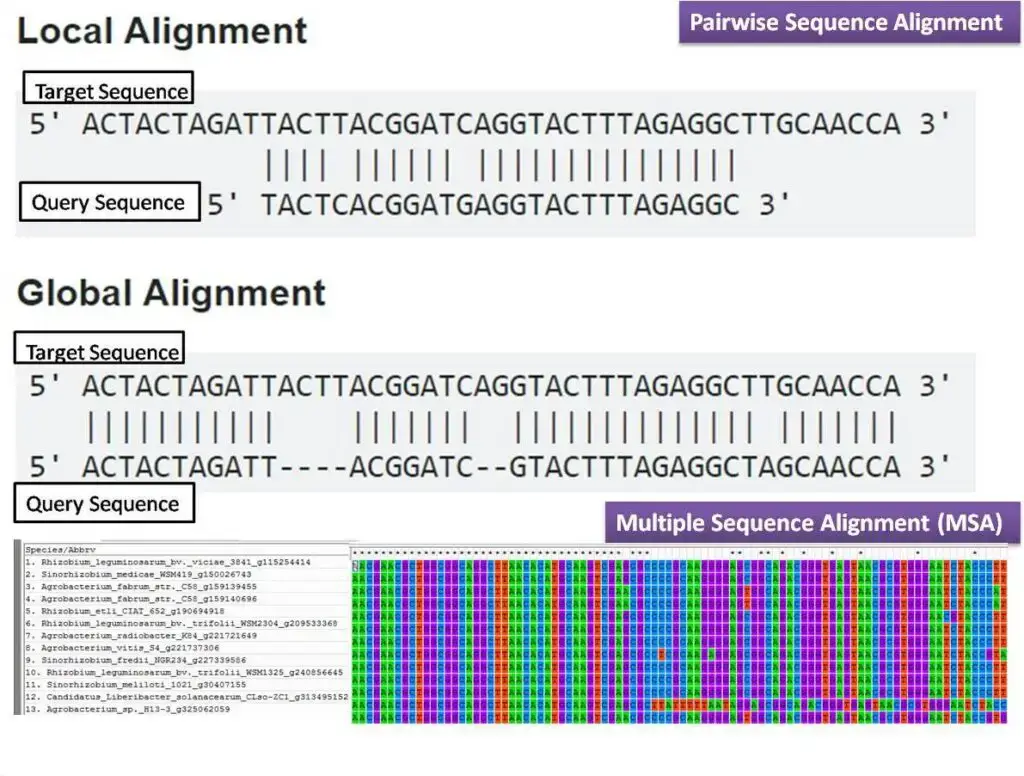
- Pairwise Alignment: Pairwise alignment is a computational technique that entails the comparison and alignment of two sequences with the aim of identifying their similarities and dissimilarities. The objective is to ascertain the optimal arrangement of sequences with a view to maximising matches while minimising mismatches and indels. The two commonly used algorithms for pairwise alignment are the Needleman-Wunsch algorithm, which is based on dynamic programming and is used for global alignment, and the Smith-Waterman algorithm, which is used for local alignment. The technique of global alignment involves the comparison of the complete length of two sequences, whereas local alignment is centred on the detection of particular regions of similarity present within the sequences.
- Multiple Sequence Alignment (MSA): The process of aligning three or more sequences simultaneously is known as Multiple Sequence Alignment (MSA). The MSA methodology expands upon pairwise alignment by integrating supplementary sequences to unveil conserved regions and evolutionary connections across a multitude of sequences. Comparing related sequences from different species or identifying common structural and functional motifs is a particularly valuable approach. The algorithms utilised in Multiple Sequence Alignment (MSA) can be broadly classified into two categories: progressive methods and iterative methods. ClustalW and T-Coffee are examples of progressive methods utilised in sequence alignment. These methods progressively construct the alignment by initially aligning pairs of sequences and subsequently integrating additional sequences. Iterative techniques, exemplified by MUSCLE and MAFFT, iteratively enhance the alignment by aligning subsets of sequences and revising the alignment based on the initial outcomes.
Pairwise alignment and multiple sequence alignment (MSA) are fundamental techniques in the field of bioinformatics. These methods enable scholars to scrutinise genetic and protein sequences, explore evolutionary connections, detect conserved regions, and forecast functional components. The selection of the alignment technique is contingent upon the particular research inquiry, the quantity of sequences under comparison, and the intended degree of sensitivity and precision.
Methods of pairwise sequence alignment
Various techniques exist for aligning sequences in pairs, such as:
- Dynamic Programming: Dynamic programming is a popular approach for global pairwise sequence alignment, with the Needleman-Wunsch algorithm being a prominent example. The algorithm generates an alignment matrix through a stepwise process of assigning scores to every conceivable alignment of pairs of subsequences. Subsequently, the matrix is employed to retrace the steps and ascertain the most advantageous alignment with the maximum score.
- Smith-Waterman Algorithm: The Smith-Waterman algorithm is a frequently utilised method for conducting local pairwise sequence alignment. The algorithm in question bears resemblance to the Needleman-Wunsch algorithm, albeit with the added capability of accommodating local alignments through the treatment of negative scores as null values. The algorithm in question employs an iterative approach to identify the local alignment that yields the highest score. This is achieved by progressively populating scores and subsequently backtracking from the position that yields the highest score.
- BLAST (Basic Local Alignment Search Tool): The Basic Local Alignment Search Tool (BLAST) is a heuristic algorithm that is commonly employed for swift pairwise sequence alignment. The tool conducts a search of a database in order to identify local alignments that exhibit a high degree of similarity to a given query sequence. The BLAST methodology employs a rapid and effective computational algorithm that concentrates on identifying noteworthy matches through the identification of high-scoring segment pairs (HSPs). Comparing large databases of sequences is especially advantageous.
- FASTA (Fast All-At-Once Sequence Comparison): The FASTA algorithm, known as Fast All-At-Once Sequence Comparison, is a commonly employed method for conducting pairwise sequence alignment. The methodology employed involves a heuristic algorithm to locate proximate similarities among sequences. The FASTA algorithm employs a dynamic programming-based approach to identify high-scoring alignments by initially searching for short word matches between the two sequences. This method offers a rapid and highly responsive approach to comparing sequences.
- Dot Plot: The dot plot is a graphical technique employed to represent pairwise sequence alignments. The process entails the representation of a sequence on the horizontal axis and another sequence on the vertical axis. Every point on the graph corresponds to a set of aligned residues, and dots are situated at the locations where the residues exhibit similarity. Dot plots offer a rapid and concise graphical representation of the resemblances and distinctions among sequences.
The aforementioned techniques exhibit differences with respect to their computational intricacy, responsiveness, and velocity. The selection of a pairwise alignment technique is contingent upon various factors, including but not limited to the length of the sequences, the desired degree of sensitivity, the computational resources at hand, and the particular research goals.
Methods of Multiple Sequence Alignment
Multiple Sequence Alignment (MSA) is a more complex task compared to pairwise alignment, as it involves aligning three or more sequences simultaneously. Several methods have been developed for MSA, including:
- Progressive Methods: Progressive methods are commonly used for MSA. These algorithms build the alignment progressively by initially aligning pairs of sequences and then incorporating additional sequences one by one. The alignment is constructed in a hierarchical manner, using a guide tree that represents the evolutionary relationships between the sequences. Popular progressive methods include ClustalW, Clustal Omega, and T-Coffee.
- Iterative Methods: Iterative methods, also known as iterative refinement methods, improve the alignment iteratively by refining an initial alignment. These algorithms typically involve three steps: (a) generating an initial alignment using a pairwise alignment algorithm, (b) estimating a new alignment based on the initial alignment, and (c) repeating the process until convergence. Common iterative methods include MUSCLE (Multiple Sequence Comparison by Log-Expectation), MAFFT (Multiple Alignment using Fast Fourier Transform), and ProbCons (Probability-based Consistency).
- Hidden Markov Model (HMM)-based Methods: HMM-based methods use probabilistic models, known as Hidden Markov Models, to align multiple sequences. These algorithms construct a statistical model that represents the conservation and variation of residues across the sequences. Popular HMM-based methods include HMMER and SAM (Statistical Alignment Model).
- Consensus-based Methods: Consensus-based methods aim to find a consensus sequence that represents the most likely alignment of the input sequences. These algorithms consider both pairwise and multiple alignments to identify the most conserved regions and common patterns across the sequences. Consensus-based methods are often used in conjunction with other alignment algorithms.
- Progressive-Iterative Methods: Progressive-iterative methods combine the advantages of both progressive and iterative approaches. They start with progressive alignment to build an initial alignment and then refine it iteratively. These methods attempt to strike a balance between speed and accuracy. Examples of progressive-iterative methods include POA (Partial Order Alignment) and DIALIGN.
Each MSA method has its own strengths, limitations, and computational requirements. The choice of method depends on factors such as the number and length of sequences, the desired alignment quality, the available computational resources, and the specific research goals. It is often recommended to compare and evaluate the results obtained from multiple alignment methods to ensure the robustness of the alignment.
Sequence alignment tool
Numerous sequence alignment tools are currently in widespread use, which aid in the process of pairwise and multiple sequence alignment. The following are commonly utilized instruments:
Tools for Pairwise Alignment:
- BLAST (Basic Local Alignment Search Tool): The Basic Local Alignment Search Tool (BLAST) is a frequently employed software application utilized for expeditious pairwise sequence alignment. The tool offers diverse search options, such as BLASTN, BLASTP, BLASTX, and others, and is equipped with the ability to perform alignments for both nucleotide and protein sequences. The National Center for Biotechnology Information (NCBI) BLAST platform, accessible at https://blast.ncbi.nlm.nih.gov/, offers a user-friendly interface for conducting BLAST inquiries.
- EMBOSS Needle: Needle is a tool for pairwise sequence alignment that is made available through the EMBOSS (European Molecular Biology Open Software Suite) package. The Needleman-Wunsch algorithm is utilized for conducting global alignment, and the tool is accessible as a standalone command-line application or via multiple online interfaces.
- EMBOSS Water: The EMBOSS package offers a pairwise alignment tool called Water, which utilizes the Smith-Waterman algorithm to conduct local sequence alignment. The tool in question is capable of identifying local similarity regions between sequences and is accessible through both standalone software and online interfaces.
Multiple Sequence Alignment Tools:
- ClustalW and Clustal Omega: ClustalW and its successor, Clustal Omega, are commonly employed progressive algorithms for multiple sequence alignment. The progressive alignment approach is utilized by them and they are accessible in the form of standalone programs, web servers, and command-line tools. The Clustal Omega software is recognized for its capacity to effectively manage extensive sequence alignments and its scalability.
- MAFFT (Multiple Alignment using Fast Fourier Transform): The MAFFT tool is an iterative approach to multiple sequence alignment that employs a variety of algorithms, such as FFT, to achieve precise and rapid alignments. The software presents alternatives for the alignment of nucleotide and protein sequences and proposes diverse tactics, including the L-INS-i, G-INS-i, and E-INS-i approaches, to suit different alignment circumstances.
- MUSCLE (Multiple Sequence Comparison by Log-Expectation): MUSCLE, which stands for Multiple Sequence Comparison by Log-Expectation, is a computational tool used for aligning multiple biological sequences. MUSCLE is a frequently employed software application for conducting multiple sequence alignment. The employed algorithm is both rapid and effective in producing precise alignments. The MUSCLE algorithm is capable of processing alignments on a large scale and provides users with various options to enhance alignment refinement and accuracy.
- T-Coffee: T-Coffee is a flexible tool for aligning multiple sequences, which utilizes a guide tree to construct alignments by integrating data from various methods. The acronym T-Coffee stands for Tree-based Consistency Objective Function for alignment Evaluation. The software incorporates multiple alignment algorithms to generate precise alignments and offers supplementary functionalities, such as predictions of secondary structures and functional domains.
The above examples of sequence alignment tools are merely a subset of the numerous of specialized tools and software that cater to specific alignment objectives or are customized to meet particular research requirements. The selection of a tool is contingent upon various factors, including but not limited to the nature of the sequences, the preferred alignment methodology, the desired functionalities, and the computational resources at hand.
Sequence alignment softwares download link
There are several widely used sequence alignment software programs available for pairwise and multiple sequence alignment. Here are some popular ones:
Pairwise Alignment Software:
- NCBI BLAST (Basic Local Alignment Search Tool): BLAST is a widely used software suite for rapid pairwise sequence alignment. It provides various programs, such as BLASTN, BLASTP, and BLASTX, for nucleotide and protein sequence alignments. BLAST can be run through the command line or accessed through the NCBI BLAST website (https://blast.ncbi.nlm.nih.gov/).
- EMBOSS (European Molecular Biology Open Software Suite): EMBOSS is a comprehensive software suite that includes multiple tools for sequence alignment, including Needle and Water for pairwise alignments. EMBOSS provides a wide range of bioinformatics tools for sequence analysis and can be downloaded from the EMBOSS website (http://emboss.sourceforge.net/).
- FASTA: FASTA is a suite of programs for pairwise sequence alignment and database searching. The package includes tools like FASTA36 and TFASTX for protein sequence alignment and FASTA3 for nucleotide sequence alignment. FASTA can be obtained from the FASTA website (https://fasta.bioch.virginia.edu/fasta_www2/fasta_list2.shtml).
Multiple Sequence Alignment Software:
- Clustal Omega: Clustal Omega is a widely used software for multiple sequence alignment. It is known for its scalability and ability to handle large-scale alignments efficiently. Clustal Omega can be accessed through the EMBL-EBI web server (https://www.ebi.ac.uk/Tools/msa/clustalo/) or downloaded as a standalone program.
- MAFFT (Multiple Alignment using Fast Fourier Transform): MAFFT is a popular software for multiple sequence alignment that uses a range of algorithms to achieve high-speed and accurate alignments. It offers several methods, including L-INS-i, G-INS-i, and E-INS-i, to handle different alignment scenarios. MAFFT can be downloaded from the MAFFT website (https://mafft.cbrc.jp/alignment/software/) or accessed through web servers.
- MUSCLE (Multiple Sequence Comparison by Log-Expectation): MUSCLE is a widely used software for multiple sequence alignment that employs an efficient algorithm to generate accurate alignments. It can handle large-scale alignments and offers options for refining alignments and improving accuracy. MUSCLE can be downloaded from the MUSCLE website (https://www.drive5.com/muscle/) or accessed through web servers.
- T-Coffee: T-Coffee is a versatile software for multiple sequence alignment that combines information from multiple methods and builds alignments based on a guide tree. It integrates various alignment algorithms to produce accurate alignments and provides additional features, such as secondary structure and functional domain predictions. T-Coffee can be downloaded from the T-Coffee website (http://www.tcoffee.org/) or accessed through web servers.
What is muscle sequence alignment?
MUSCLE (Multiple Sequence Comparison by Log-Expectation) is a popular software program used for multiple sequence alignment (MSA). It is designed to efficiently align three or more sequences simultaneously, aiming to generate accurate alignments for a variety of biological sequences, including nucleotide and protein sequences.
MUSCLE utilizes a progressive alignment strategy, which starts by building a guide tree based on pairwise sequence similarities. It then aligns sequences progressively, adding one sequence at a time to the growing alignment. The process involves several stages:
- Creating a Distance Matrix: MUSCLE calculates the pairwise distances between sequences and constructs a distance matrix. The distance measure used is typically the number of observed substitutions per site.
- Building a Guide Tree: The distance matrix is used to build a guide tree, which represents the evolutionary relationships between the sequences. The guide tree serves as a roadmap for aligning the sequences in a hierarchical manner.
- Progressive Alignment: MUSCLE performs the alignment progressively by aligning pairs of sequences based on the guide tree. Starting from the most closely related sequences, MUSCLE aligns them and extends the alignment to include the next sequence in the guide tree, iteratively building the alignment.
- Iterative Refinement: MUSCLE employs an iterative refinement process to improve the alignment. It iteratively adjusts the alignment by optimizing an objective function that takes into account both sequence similarity and consistency with the guide tree. This refinement step aims to enhance the accuracy of the alignment.
MUSCLE is known for its speed and scalability, capable of handling large-scale sequence alignments efficiently. It offers various options to customize the alignment process, such as adjusting the alignment quality, choosing different distance measures, and incorporating user-specified constraints.
MUSCLE can be used as a standalone program that runs from the command line or as a web-based tool through servers provided by the developers or other online platforms. It has been widely adopted in bioinformatics and is valuable for various applications, including comparative genomics, phylogenetic analysis, protein structure prediction, and functional annotation.
Aequence alignment algorithm
Sequence alignment algorithms are computational methods used to align two or more biological sequences, such as nucleotide or protein sequences, to identify regions of similarity or homology. These algorithms aim to determine the optimal alignment by assigning scores to different alignment possibilities and finding the alignment with the highest score. Here are some commonly used sequence alignment algorithms:
- Dynamic Programming Algorithms:
- Needleman-Wunsch Algorithm: The Needleman-Wunsch algorithm is a dynamic programming algorithm used for global pairwise sequence alignment. It constructs an alignment matrix by iteratively filling in scores for all possible alignments of subsequence pairs. The alignment with the highest score is determined by backtracking through the matrix.
- Smith-Waterman Algorithm: The Smith-Waterman algorithm is a dynamic programming algorithm used for local pairwise sequence alignment. It is similar to the Needleman-Wunsch algorithm but allows for local alignments by considering negative scores as zero. The algorithm identifies the best-scoring local alignment by iteratively filling in scores and backtracking from the highest-scoring position.
- Heuristic Algorithms:
- BLAST (Basic Local Alignment Search Tool): BLAST is a widely used heuristic algorithm for pairwise sequence alignment. It performs a database search to find local alignments that are highly similar to a query sequence. BLAST utilizes a fast and efficient algorithm that focuses on high-scoring segment pairs (HSPs) to identify significant matches.
- FASTA (Fast All-At-Once Sequence Comparison): FASTA is a heuristic algorithm for pairwise sequence alignment. It scans both sequences for short word matches and extends them into high-scoring alignments using a dynamic programming-based algorithm. FASTA provides a fast and sensitive method for sequence comparison.
- Progressive Alignment Algorithms:
- Progressive alignment algorithms are used for multiple sequence alignment (MSA) and follow a step-by-step process to align sequences progressively.
- The progressive alignment method starts with pairwise alignments, constructs a guide tree based on the similarity between sequences, and then aligns sequences in a hierarchical manner using the guide tree as a reference. Popular progressive alignment tools include ClustalW and Clustal Omega.
- Iterative Refinement Algorithms:
- Iterative refinement algorithms are used for improving alignments iteratively to increase accuracy.
- These algorithms typically involve an initial alignment step, followed by iterations of refining the alignment based on scoring functions and optimization techniques. Examples include the iterative refinement step in MUSCLE (Multiple Sequence Comparison by Log-Expectation) and the ProbCons algorithm.
Applications of sequence alignment
Sequence alignment has a wide range of applications in various fields, including:
- Genomics: Sequence alignment plays a crucial role in genome assembly, where it helps in piecing together short DNA fragments into complete genomes. It is also essential for identifying genes, regulatory regions, and functional elements within genomes. Comparative genomics uses sequence alignment to study evolutionary relationships between species and identify conserved regions.
- Proteomics: Sequence alignment is used to compare protein sequences and identify similarities, functional domains, and motifs. It aids in predicting protein structure and function, as well as understanding the relationship between protein sequences and their biological activities. It is also instrumental in protein family classification and drug target identification.
- Evolutionary Biology: Sequence alignment allows researchers to study the evolutionary history of species by comparing DNA or protein sequences. It helps in inferring phylogenetic trees and determining the relatedness between different organisms. By aligning sequences from different species, scientists can trace the origin and divergence of genetic material.
- Drug Discovery: Sequence alignment assists in identifying potential drug targets by comparing protein sequences of disease-related genes. It helps in understanding the functional implications of genetic variations and mutations associated with diseases. Sequence alignment is also used in virtual screening, where it aids in identifying drug candidates by aligning them with target protein sequences.
- Forensic Analysis: Sequence alignment is employed in forensic DNA analysis, where it helps in comparing DNA profiles obtained from crime scenes with those of potential suspects. By aligning DNA sequences, scientists can determine matches or mismatches, providing evidence for identification or exclusion.
- Molecular Biology and Biotechnology: Sequence alignment aids in designing primers for polymerase chain reaction (PCR) experiments, where specific regions of DNA are amplified. It is also used in recombinant DNA technology to align DNA sequences for the purpose of gene cloning, genetic engineering, and gene synthesis.
- Biodiversity and Conservation: Sequence alignment enables researchers to study genetic diversity within and between species, contributing to biodiversity assessments and conservation efforts. It helps in identifying unique genetic markers, assessing population structure, and understanding the genetic basis of adaptation and speciation.
- Personalized Medicine: Sequence alignment is utilized in personalized medicine to analyze an individual’s genetic variation and identify disease-causing mutations or genetic predispositions. It aids in tailoring treatment strategies and predicting drug responses based on an individual’s genetic profile.
These are just a few examples of the numerous applications of sequence alignment. It is a versatile tool that provides insights into the structure, function, evolution, and relationship of biological sequences, contributing to advancements in various scientific disciplines and practical applications.
FAQ
What is sequence alignment, and why is it important?
Sequence alignment is the process of arranging and comparing two or more biological sequences to identify regions of similarity or homology. It is important because it helps researchers understand the relationships between sequences, identify conserved regions, detect functional domains, predict protein structure and function, and infer evolutionary relationships.
How does the Needleman-Wunsch algorithm work for global sequence alignment?
The Needleman-Wunsch algorithm is a dynamic programming algorithm for global sequence alignment. It constructs an alignment matrix by assigning scores to all possible alignments of subsequence pairs. The alignment with the highest score is determined by backtracking through the matrix, providing the optimal alignment.
What is the Smith-Waterman algorithm, and how does it perform local sequence alignment?
The Smith-Waterman algorithm is a dynamic programming algorithm for local sequence alignment. It is similar to the Needleman-Wunsch algorithm but allows for negative scores to be treated as zero. The algorithm identifies the best-scoring local alignment by iteratively filling in scores and backtracking from the highest-scoring position.
What are the different types of sequence alignment methods?
The two main types of sequence alignment methods are global alignment and local alignment. Global alignment compares the entire length of sequences, while local alignment focuses on aligning specific regions of similarity. Other methods include pairwise alignment for comparing two sequences and multiple sequence alignment for aligning three or more sequences.
Can you explain the concept of scoring matrices in sequence alignment?
Scoring matrices are used in sequence alignment to assign scores to different alignment possibilities. They quantify the similarity or dissimilarity between residues based on their occurrence frequencies in a set of aligned sequences. The most commonly used scoring matrix is the BLOSUM (Blocks Substitution Matrix) series for protein sequences.
What are some popular software tools for pairwise sequence alignment?
Some popular software tools for pairwise sequence alignment include BLAST (Basic Local Alignment Search Tool), EMBOSS Needle, and FASTA. These tools provide user-friendly interfaces or command-line options to perform pairwise alignments, allowing researchers to analyze and compare sequences efficiently.
How does multiple sequence alignment differ from pairwise alignment?
Multiple sequence alignment (MSA) aligns three or more sequences simultaneously, while pairwise alignment compares only two sequences. MSA takes into account the relationships between multiple sequences to identify conserved regions, insertions, and deletions. It is useful for studying evolutionary relationships and identifying functional elements.
Which algorithm is commonly used for multiple sequence alignment?
Progressive alignment algorithms, such as ClustalW, Clustal Omega, MAFFT, and MUSCLE, are commonly used for multiple sequence alignment. These algorithms build a guide tree based on pairwise sequence similarities and align sequences progressively based on the tree, producing accurate alignments.
What are some applications of sequence alignment in genomics and proteomics?
Sequence alignment has numerous applications in genomics and proteomics. It is used for genome assembly, identifying gene sequences, annotating genetic variants, predicting protein structure and function, studying evolutionary relationships, and designing primers for PCR amplification, among many other applications.
How do I interpret the output of a sequence alignment, including gap penalties and alignment scores?
Gap penalties represent the cost or penalty assigned for introducing gaps (insertions or deletions) in the alignment. Higher gap penalties discourage the introduction of gaps, favoring more conserved alignments. Alignment scores indicate the overall similarity between the aligned sequences, with higher scores indicating greater similarity.
References
- Needleman, S.B., and Wunsch, C.D. (1970). A general method applicable to the search for similarities in the amino acid sequence of two proteins. Journal of Molecular Biology, 48(3), 443-453.
- Smith, T.F., and Waterman, M.S. (1981). Identification of common molecular subsequences. Journal of Molecular Biology, 147(1), 195-197.
- Altschul, S.F., et al. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403-410.
- Thompson, J.D., et al. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673-4680.
- Katoh, K., et al. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Research, 30(14), 3059-3066.
- Edgar, R.C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32(5), 1792-1797.
- Notredame, C., et al. (2000). T-Coffee: a novel method for fast and accurate multiple sequence alignment. Journal of Molecular Biology, 302(1), 205-217.
- Larkin, M.A., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947-2948.
- Sievers, F., and Higgins, D.G. (2014). Clustal Omega for making accurate alignments of many protein sequences. Protein Science, 27(1), 135-145.
- Katoh, K., et al. (2019). MAFFT version 7: improvements in performance and usability. Molecular Biology and Evolution, 30(4), 772-780.