What is Nitrogen Fixation?
Nitrogen fixation is the fundamental biological process and the first step in the nitrogen cycle. Certain bacterial species, such as Rhizobium, Azotobacter, etc., and other natural phenomena convert atmospheric nitrogen into ammonia (another type of nitrogen) throughout this process.
- NASA reports that the air in the earth’s atmosphere is composed of 78% nitrogen gas, 21% oxygen, 0.93 % argon, 0.04 % carbon dioxide, and trace amounts of other gases. Numerous biological components also include nitrogen.
- The concentration of an useable form of nitrogen is extremely low despite everything.
- Nitrogen gas is a necessary yet scarce ingredient.
- Nitrogen is required for the cellular functions of all species, including humans, plants, and other organisms.
- Nitrogen fixation is a chemical process in which molecular nitrogen (N2) in the air is transformed into ammonia (NH3) or related nitrogenous chemicals, mainly in soil or aquatic environments but also in industry.
- Aside from a few microbes, atmospheric nitrogen is molecular dinitrogen, a generally nonreactive chemical that is biologically inert. Biological nitrogen fixation, also known as diazotroph, is a crucial microbially-mediated activity that transforms dinitrogen (N2) gas to ammonia (NH3) via the nitrogenase protein complex (Nif).
- Nitrogen fixation is required for the creation of all nitrogen-containing organic molecules, including amino acids, proteins, nucleoside triphosphates, and nucleic acids.
- As a component of the nitrogen cycle, it is crucial for agriculture and fertiliser production.
- Indirectly, it is also relevant to the production of all nitrogen-based chemical products, including certain explosives, medicines, and colours.
- Diazotrophic microorganisms, which include bacteria such as Azotobacter and archaea, are responsible for nitrogen fixation in the soil.
- There are symbiotic interactions between certain nitrogen-fixing bacteria and plant groupings, particularly legumes.
- As demonstrated by nitrogen fixation on rice roots, looser non-symbiotic connections between diazotrophs and plants are generally referred to as associative.
- There is nitrogen fixation between certain termites and fungus. It occurs naturally in the atmosphere because lightning produces NOx.
- Enzymes known as nitrogenases catalyse all biological events involved the nitrogen fixation process.
- These enzymes contain iron and frequently a second metal, most commonly molybdenum but also vanadium.
Why is Nitrogen Fixation Necessary?
It serves the following functions in nature:
- Makes nitrogen accessible for the growth and metabolism of plants and animals.
- Contributes to the continuation of the nitrogen cycle in nature, where nitrogen fixation is a key stage.
- Synthetically produces nitrogenous fertilisers and insecticides on a big scale. With horticulture, this aids in crop production and increases their output.
- Assists in the formation of our body’s structural macromolecules, such as proteins and nucleic acids (RNA and DNA).
Nitrogen Fixation Types
- Nitrogen is the most abundant necessary macroelement among carbon, hydrogen, and oxygen. Nitrogen is necessary for the synthesis of amino acids, proteins, nucleic acids, cytochromes, chlorophylls, alkaloids, phytohormones, and many vitamins in plants.
- Microorganisms and plants struggle for the limited nitrogen content in the soil. Plants primarily absorb nitrogen from the soil in the form of nitrate (NO3–) or ammonium ions (NH4+).
- Nitrate is more prevalent in well-oxygenated, non-acidic soils, whereas ammonium is more prevalent in acidic or waterlogged soils.
- Other sources of available soil nitrogen may include amino acids from decaying organic matter, animal excrement (urea), and plant-available artificial fertilisers.
- Plants acquire nitrogen primarily from the atmosphere. It exists as diatomic molecules (N2) in the air. Highly inert gas is present.
- It cannot be immediately utilised by the higher plants and must thus be fixed.
- Nitrogen fixation is the conversion of free nitrogen (molecular and elemental) into nitrogenous compounds (to make it available for absorption by plants).
- The fixation of nitrogen is accomplished through physicochemical and biological processes. About 10% of natural nitrogen fixation is carried out by physicochemical processes, whereas the remaining 90% is carried out by biological processes.
- Physical nitrogen fixation and biological nitrogen fixation are the two types of nitrogen fixation.
1. Physical Nitrogen Fixation
(i) Natural Nitrogen Fixation
Under the influence of lightning (i.e., electric discharge in the clouds) and thunder, the air’s N2 and O2 combine to generate nitric oxide (NO). Nitric oxides are reoxidized with oxygen to produce nitrogen peroxide (NO2).
The following are the responses:
N2 + O2 Lightning → Thunder 2N0 (Nitric Oxide); 2NO + O2 → 2NO2 Oxidation (Nitrogen peroxide)
NO2 mixes with rainwater to generate nitrous acid (HNO2) and nitric acid during precipitation (HNO3). Acids combine with alkaline radicals to generate water-soluble nitrates (NO3-) and nitrites when they fall to the ground with rainwater (N02-).
2NO2 + H2O → HNO2 + HNO3; HNO3 + Ca or K salts → Ca or K nitrates
The nitrates are water-soluble and are directly absorbed by the plant roots.
(ii) Industrial Nitrogen Fixation
Ammonia is manufactured through the direct combination of nitrogen and hydrogen (obtained from water) under high temperature and pressure. Later, it is turned into a variety of fertilisers, including urea and others.
2. Industrial Methods of Nitrogen Fixation
a. Haber or Haber-Bosch process
- The most prevalent commercial technology for nitrogen fixation and ammonia production is the Haber or Haber-Bosch process.
- Fritz Haber described the process, which earned him the 1918 Nobel Prize in Chemistry, and Karl Bosch adapted it for commercial usage in the early 20th century.
- Ammonia is produced by heating and pressurising nitrogen and hydrogen in a tank containing an iron catalyst.
b. Cyanamide process
- The cyanamide process generates calcium cyanamide (CaCN2, commonly known as Nitrolime) by heating calcium carbide in an environment of pure nitrogen.
- As a result, calcium cyanamide is utilised as a plant fertiliser.
c. Electric arc process
- In 1895, Lord Rayleigh invented the electric arc process, the first synthetic method for fixing nitrogen.
- Similar to how lightning fixes nitrogen in nature, the electric arc technique fixes nitrogen in a laboratory.
- By combining oxygen and nitrogen in the air, an electric arc produces nitrogen oxides. Air containing oxides is bubbled through water to produce nitric acid.
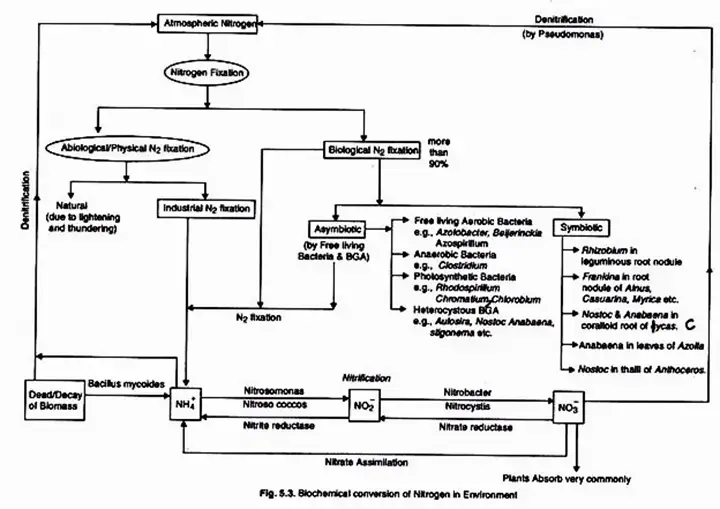
3. Biological Nitrogen Fixation
Biological nitrogen fixation refers to the transformation of atmospheric nitrogen into nitrogenous compounds through the intervention of living organisms. Two basic types of microorganisms carry out the process: those that live in tight symbiotic relationship with other plants and those that are “free dwelling” or non-symbiotic.
BNF is the process by which atmospheric nitrogen is converted into ammonia in the presence of nitrogenize. Nitrogenize is a biological catalyst exclusive to particular microbes, such as the symbiotic Rhizobium and Frankia or the free-living Azospirillum, Azotobacter, and BGA.
Following are specifics on biological nitrogen fixing.
Nearly 80% of the Earth’s atmosphere is composed of nitrogen in the form of a very inert di-nitrogen (N = N), which most plants are unable to exploit. The di-nitrogen (N2) in the atmosphere consists of two nitrogen atoms connected by a triple-covalent bond. Approximately 225 kcal of energy are required to break this tough triple bond.
Biological nitrogen fixation, also known as diazotrophy, is the transformation of inert gaseous di-nitrogen (N2) into plant-available ammonia (NH3) via the action of microorganisms.
Nitrogen Fixers
Only certain prokaryotes, such as bacteria and cyanobacteria, are capable of fixing atmospheric nitrogen. They are known as nitrogen-fixing bacteria or diazotrophs. By natural mechanism, they fix around 95% of the total global nitrogen fixed annually (-200 million metric tonnes). Diazotrophs may be asymbiotic (free-living) or symbiotic, as described in the table below.
(i) Free Living Nitrogen Fixing Bacteria
- Nitrogen fixing is performed by the saprophytic bacteria Azotobacter, Beijerinckia (bothaerobic), and Clostridium (anaerobic).
- Desulphovibrio is a chemotrophic bacteria that fixes nitrogen. Rhodopseudomonas, Rhodospirillum, and Chromatium are photoautotrophic bacteria that fix nitrogen.
- These bacteria add 10 to 25 kg of nitrogen per hectare each year.
(ii) Free living Nitrogen Fixing Cyanobacteria
- Numerous free-living blue-green algae (today known as cyanobacteria) fix nitrogen, including Anabaena, Nustoc, Aulosira, Cylmdrospermum, and Trichodesmium.
- These are ecologically significant because they inhabit waterlogged soils where denitrifying bacteria can be active.
- The most active nitrogen fixer in rice fields is Aulosira fertilissima, whereas Cylindrospermum is active in sugarcane and maize fields. They add 20-30 kg of nitrogen per hectare each year.
(iii) Symbiotic Nitrogen Fixing Cyanobacteria
- Anabaena and Nostoc species are widespread symbionts in the roots of lichens, Anthoceros, Azolla, and cycads.
- The water fern Azolla pinnata has Anabaena azollae in its fronds. It is frequently introduced to rice fields to fix nitrogen.
(iv) Symbiotic Nitrogen Fixing Bacteria
- Rhizobium are gram-negative, aerobic, nitrogen-fixing symbionts of Papilionaceous roots.
- Root nodules of Sesbania rostrata contain Rhizobium, while stem nodules contain Aerorhizobium. Frankia is found in the root nodules of numerous non-leguminous plants, such as Casuarina and Alnus.
- Xanthomonas and Mycobacterium are symbionts in the leaves of certain Rubiaceae and Myrsinaceae species (e.g., Ardisia).
- Several soil-dwelling Rhizobium species are incapable of fixing nitrogen on their own. They only do so as symbionts associated with the roots of legumes.
- The symbiotic nitrogen fixation can be broken down into the subsequent steps:
(i) Nodule formation
It requires several interactions between soil-dwelling Rizobium and the host plant’s roots. The significant phases in nodule development are as follows: -Host Particularity: A variety of microorganisms inhabit the rhizosphere (i.e., the region surrounding the roots) of host roots.
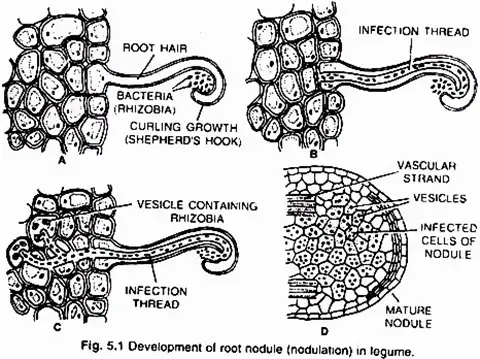
Young leguminous plants exude a set of chemical attractants, including flavonoids and betaines, from their roots. Specific rhizobial Tells migrate near root hairs in response to these chemical attractants and create nod (nodulation) factors. The nod factors on the surface of bacteria bind to the lectin proteins on the surface of root hairs. This contact stimulates the development and curling of root hairs surrounding Rhizobia.
At these sites, the cell wall breaks in response to node-factors, allowing Rhizobia to enter the root hair by an invagination of plasma membrane known as an infection thread. The infection thread containing dividing Rhizobia extends through the root hair before branching out to reach various cortical cells.
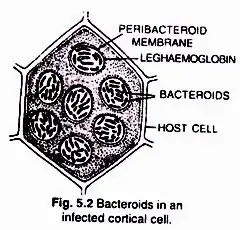
Rhizobia are discharged into the cortical cells either singly or in membrane-enclosed groups. The Rhizobia cease to divide, lose their cell wall, and transform into nitrogen-fixing cells known as lead bacteroids. The membrane that surrounds bacteroids is known as the peribacteroid membrane. Infected cortical cells produce nodules by dividing.
(ii) Mechanism of nitrogen fixation
The nodule serves as a N2 fixation site. It contains all important biochemicals, such as the nitrogenase enzyme complex and leghaemoglobin (leguminous haemoglobin). Mo-Fe protein (molybdoferredoxin) and Fe-protein are the two components of nitrogenase (azoferredoxin). The nitrogenase catalyses the conversion of di-nitrogen (N2) in the environment to 2NH3. The first stable product of nitrogen fixation is ammonia.
The overall formula is:
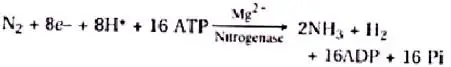
The nitrogenase is particularly oxygen-sensitive. In order to safeguard these enzymes, nodule includes a reddish-pink oxygen scavenger called leghaemoglobin (Lb). There are two perspectives about the location of leghaemoglobin, which is either outside the peribacteroid membrane or between bacteroids.
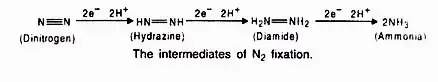
During nitrogen fixation, the free di-nitrogen binds to the MoFe protein and is not released until the ammonia is entirely reduced. Numerous intermediates are produced during the reduction of di-nitrogen to yield ammonia (NH3), which is protonated at physiological pH to form NH4+. In this process, ferredoxin acts as an electron donor to Fe-protein (nitrogenase reductase), which hydrolyzes ATP to reduce MoFe protein, which then reduces the substrate N2. Photosynthesis and respiration of the host cells provide the electrons and ATP.
Assimilation of Ammonia:
Nitrogenase produces ammonia, which is instantly protonated to form ammonium ion (NH4+). As NH4+ is poisonous to plants, it is swiftly converted into amino acids at its point of generation. The synthesis of amino acids involves three processes: reductive animation, catalytic amination, and transamination.
(i) Reductive amination
Glumate dehydrogenase (GDH) catalyses the synthesis of glutamic acid in this mechanism.
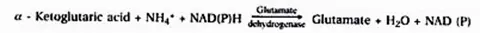
(ii) Catalytic amidation
It involves two enzymes, glutamine synthetase (GS) and glutamate synthetase (glutamine – 2-oxyglutarate aminotransferase, or GOGAT).

One of the two generated glutamates is returned to GS, while the other is sent to the factory.
(iii) Transamination
From glutamate or glutamic acid, other amino acids are produced via transamination. Aminotransferases (= transaminases) are the enzymes that catalyse all of these processes. In transamination, the amino group of one amino acid is transferred to the keto group of another amino acid.
Glutamate (amino donor) + Oxaloacetate (amino acceptor) → Aspartate (amino acid) + 2 oxyglutarate
In nitrogen-fixing plants, the fixed nitrogen is exported from the nodules to other plant parts via xylem in the form of amides (asparagines and glutamine) and Ureides (allantoin, allantoic acid, and citrulline). Amides are generated by replacing the – OH portion of glutamic acid and aspartic acid with another NH2– radicle. Consequently, amides have more nitrogen than amino acids and are structural components of the majority of proteins.
Nitrate Assimilation
Nitrate cannot be used by plants in its natural state. Prior to incorporation into organic molecules, it is first converted to ammonia. The reduction of nitrate involves two steps:
1. Reduction of nitrate to nitrite
It is carried out by nitrate reductase, an inducible enzyme. It is a molybdoflavoprotein enzyme. For its function, it requires the reduced coenzymes NADH or NADPH, which are brought into contact with nitrate by FAD or FMN.
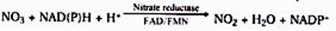
2. Reduction of nitrate:
This process is performed by the enzyme nitrite reductase. The enzyme is a metalloflavoprotein containing iron and copper. It is found in leaf chloroplasts and leucoplasts of other cells. Nitrite reductase necessitates reducing ability. It consists of NADPH and NADH (NADPH in illuminated cells).
The reduction process also requires ferredoxin, which is found in green plant tissues. In non-illuminated cells of higher plants, it is believed that either nitrite is translocated to leaf cells or another electron donor (such as FAD) functions. The product of reducing nitrite to ammonia.
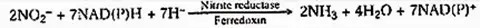
Thus generated ammonia reacts with organic acids to form amino acids. Protein is formed by the translation of amino acids.
Fixation of Atmospheric Nitrogen – Enzyme System
Fixation of atmospheric nitrogen is dependent upon the nitrogenase enzyme system, which consists of nitrogenase and nitrogenase reductase. As demonstrated in the following equation, electrons are transmitted from ferredoxin or flavodoxin to nitrogenase reductase and then to nitrogenase, where they are used to convert N2 and H+ to NH3 and H2.
N2 + 8e– + 8H+ + 16 Mg ATP → H2 + 16 Mg ADP + 16 Pi
There are additionally two co-proteins present in the nitrogenase enzyme system. These are a MoFe protein with molybdenum and iron, and a Fe protein with simply iron. The active site of nitrogenase where nitrogen reduction occurs is dependent on a cofactor containing iron and molybdenum (Fe MO CO). The generation of H2 during the reduction of nitrogen contributes to the ATP demands of nitrogen fixation. The evolution of H2 accompanies the biological fixation of nitrogen.
However, only certain strains of Rhizobium and Bradyrhizobium possess hydrogenase and can utilise hydrogen, whereas other nitrogen-fixing bacteria produce H2 gas inefficiently.
Nodulated root systems that produce hydrogen are frequently colonised by hydrogen-oxidizing Acinetobacter strains that thrive on the hydrogen produced by nitrogen-fixing bacteria.
Nitrogenase is remarkably sensitive to oxygen and is irreversibly deactivated by exposure to even low concentrations. Therefore, nitrogen fixation is typically limited to environments where nitrogenase is shielded from exposure to molecular oxygen.
Nitrogen Fixation Mechanisms in Microorganisms
1. Nitrogenase Types, Structure and Function
- The biological conversion of atmospheric nitrogen to ammonia is facilitated by the Nitrogenase enzyme.
- This enzyme is anaerobic and becomes inactive when it comes into contact with oxygen or air.
- The proteinaceous enzyme is composed of two subunits, the larger MoFe protein (molecular weight of 2,200,000 Dalton) and the smaller Fe protein (55000 Dalton mol. wt.).
- British microbiologist Postgate gave them the designations Kp1 and Kp2.
- As said, it comprises atoms of iron and molybdenum. Activation requires Mg2+ ions, and they can convert ATP to ADP during function.
- It is inhibited by ADP and reduces a number of substrates with triple-bonded molecules (similar to N = N). The enzyme can convert hydrogen ions to gaseous hydrogen, even in the presence of nitrogen dioxide, and can also convert acetylene to ethylene.
- The nitrogenase isolated from three types of bacteria exhibited the characteristics listed below. As far as the size of proteins (in both MoFe and Fe components) is concerned, this demonstrates that the nature of enzymes differs among nitrogen-fixing organisms.
(i) The Cp Type
- According to Postgate, the properties of the nitrogenase are the Cp type (Clostridium pasteurianum): It has MoFe (Cp1) and Fe (Cp2) proteins which have 2,20000 Dalton and 55,000 Dalton molecular weight, respectively. The half-life of enzymes is rather brief.
(ii) The Kp Type (Klebsiella pneumoniae)
- It contains MoFe (Kp1) and Fe (Kp2), which have molecular weights of 2,18000 and 66,700 Dalton, respectively.
(iii) The Ac Type (Azotobacter chroococcum)
- It contains MoFe (Ac1) and Fe (Ac2), which have respective molecular weights of 2,27000 and 64,000 Dalton.
- The enzyme half-lives of all three types of Fe units are significantly shorter than those of FeMo units.
2. Alternative Nitrogenase:
- Professor P. Bishop and his colleagues in the United States uncovered evidence that Azotobacter vinelandii had a distinct type of nitrogenase.
- The genomic data suggested that their nitrogenase (nif YKDH) genes had been eliminated. This nitrogenase was identified from A. chroococcum in the future. It is composed of two proteins, one large and heteromeric and the other smaller and very similar to the typical Fe-protein; both are sensitive to O2.
- The enzyme releases a single molecule of hydrogen and decreases acetylene. This ‘new’ nitrogenase incorporates vanadium in place of Mo ion in the bigger protein subunit of the conventional system.
- The fact that V-nitrogenase is suppressed by Mo suggests that it serves as a physiological “backup” nitrogen-fixing pathway for Azotobacter in the event that Mo is absent.
3. Substrates for Nitrogenase
- For enzyme activity, a substrate must bind to all of the enzyme’s active sites in order to produce a product.
- The enzymatic conversion of atmospheric nitrogen to ammonia could be described as follows.
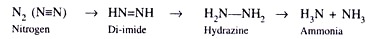
- Intriguingly, the cell-free extracts of Azotobacter and Clostridium converted nitrogen in the same manner as the free-living bacterial cells.
- This ultimately led to the isolation and purification of the enzyme from C. pasteurianum and A.chromococcum for the first time. In addition, the enzyme was in charge of the absorption and reduction of N2 gas.
- Although nitrogenase has numerous substrates, with the exception of H2, the majority of substrates are non-physiological since nitrogenase reduces the inhibitors.
- In addition, acetylene (HC=CH) is an important and reliable substrate for assessing enzyme activity using the “acetylene-reduction” assay. It is also significant to note that the majority of substrates have triple bonds identical to nitrogen (N=N) in their molecules.
The following substrates are reactive to nitrogenase:
Substrate – Products(s)
N2 – NH3
N2O – H2O, N2
N3 – NH3, N2
C2H2 – C2H4
HCN – CH4, NH3, CH3, NH2
CH3CN – C2H6, NH3
In the reduction of acetylene, ethylene is produced, which takes two electrons (HC = CH H2C-CH2), whereas the reduction of nitrogen to ammonia requires six electrons. Compared to other substrates, nitrogen can be decreased quickly.
Some of the nitrogen-fixing bacteria simultaneously create H2 when N2 is fixed. N2 fixation are related to one another. In the presence of deuterium, nitrogenase converted the H+ ion into H-D. The H-D reaction shows a bound di-imide intermediate is involved in N2 fixation.
Considering the release of H2 during nitrogen fixation, the following response has been proposed:
N2 + 8 H+ + 8e– +16 ATP → 2NH3 + H2 + 16 ADP + 16 Pi
ATP provides the energy for the nitrogenase reaction, which derives from cellular metabolic cycles. This is accomplished through photophosphorylation, oxidative phosphorylation, and phosphoroclastic dissimilation. In a subsequent step, keto-acid is converted to acetyl phosphate, carbon dioxide, and hydrogen.
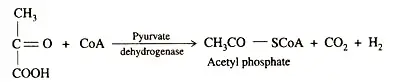
Pyruvate serves as both an electron donor and a source of energy. In the phosphoroclastic processes, pyruvate is converted to acetyl phosphate, which, when combined with ADP, produces ATP. The electron carrier proteins ferredoxin and flavodoxin are the reductants.
Dithionite (Na2S2O4) and some dyes, such as methyl viologen and benzyl viologen, can also act as extracellular electron donor sources. The transfer of electrons from pyruvate or hydrogen to ferredoxin or flavodoxin is catalysed by this enzyme system.
(a) Ferredoxins or di-nitrogenase
- Ferredoxins are electron carriers that were identified in 1962 by Mortenson and Caruahan in C. pasteurianum. It is a naturally occurring iron-sulfur (Fe-S) carrier protein (reversible).
- Currently, it has been isolated from a variety of cyanobacteria, photosynthetic bacteria, and even higher plants.
- The ferredoxins participate in numerous physiological activities, including photosynthesis in plants and pyruvate metabolism in anaerobic bacteria.
- Involved in N2 fixation, the ferredoxin molecule has one cluster of four irons and four sulphur atoms. The iron atoms of nitrogenase are composed of many similar iron-sulfur clusters. The cluster as a whole behaves as an oxido-reductive unit.
- Electron paramagnetic resonance suggests that the ferredoxins of bacteria that fix nitrogen aerobically, such as Azotobacter, behave differently than those of anaerobes, such as Clostridium pasteurianum.
- In C. pasteurianum, ferredoxin is the protein that interacts with nitrogenase and provides the reducing power necessary for the conversion of N2 to NH3.
(b) Flavodoxins or di-nitrogen reductase
- The bacteria create flavodoxin under conditions of nutritional stress, characterised by restricted iron availability. Additionally, it was initially isolated from C.pasteurianum.
- It is intriguing that it was discovered to replace ferredoxin in a wide number of processes as an electron carrier. Azotobacter vinelandii was used to isolate an electron carrier termed azotoflavin with biological activity similar to ferredoxins.
- In K.pneumoniae and A.chroococcum, flavodoxins function as the major nitrogenase reductant.
- Half-reduced A. chroococcum flavodoxin is blue while fully reduced flavodoxin is colourless. The reduced form of flavodoxin is quinone and semiquinone.
- This version is exceptionally resistant to oxidation by air, which may explain why this protein, as opposed to a ferredoxin, is more suited to Azotobacter’s aerobic lifestyle.
- Flavodoxins lack iron atoms; their oxido-reducible core is a yellow fluorescent molecule known as flavin.
The Function of the Pyruvate and Ferredoxin-Nitrogenase Reaction:
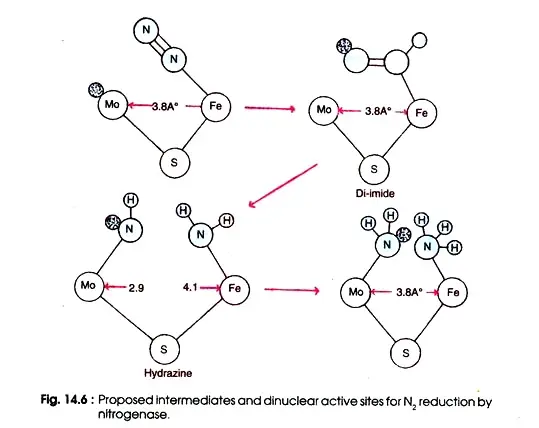
Figure depicts the enzyme’s active site for substrate reduction. This enzyme is thought to consist of a Mo-Fe di-nuclear site bridged by sulphur with the correct size and electron properties to provide a Mo-Fe distance of 3.8.
This spacing is particular in order to include various nitrogenase substrates, including nitrogen, while excluding others. The initial step in the reduction of nitrogen is the creation of a linear complex between nitrogen and the Fe atom of nitrogenase.
This is followed by the transfer of electrons from Mo, the final step of the electron activating system, resulting in the synthesis of di-imide, which is stabilised by hydrogen bonds from the protein and metal nitrogen bonds.
Hydrazine is produced by the successive addition of electrons, followed by the breaking of the N—N link to form two molecules of NH3. In order to maintain a consistent Mo—Fe distance, the increase in N—N bond length during reduction is accompanied by offsetting changes in the MNN angles.
4. Electron Proteins
MoFe (kp1) proteins play a crucial role in nitrogen fixation (substrate binding and reduction), whereas Fe protein (kp2) aids in electron transfer from flavodoxin to larger subunits and ATP consumption. neither of the two protein subunits can independently function (Fig. 14.7).
Fe proteins have four Fe centres and an equal amount of sulphur, whereas MoFe proteins have a variable number of Fe centres. It is 22-24 degrees Celsius in C.pasturianum. A comparable amount of inorganic sulphur is also present. MoFe protein, unlike Fe protein, contains two additional Mo atoms. Mo may have a crucial function in the H2 development that accompanies N2 fixation.
4. Electron Proteins
MoFe (kp1) proteins play a crucial role in nitrogen fixation (substrate binding and reduction) alongside Fe protein (kp2), which aids in electron transfer from flavodoxin and ATP consumption. neither of the two protein subunits can independently function (Fig. 14.7).
Fe proteins have four Fe centres and an equal amount of sulphur, whereas MoFe proteins have a variable number of Fe centres. It is 22-24 degrees Celsius in C.pasturianum. A comparable amount of inorganic sulphur is also present. MoFe protein, unlike Fe protein, contains two additional Mo atoms. Mo may have a crucial function in the H2 development that accompanies N2 fixation.
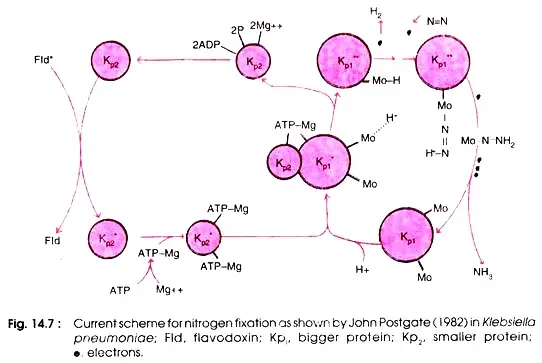
The N2 reduction process begins with the transfer of a single electron from flavodoxin to the smaller subunit of Fe protein, Kp2. At this stage, two ATP molecules combine with two ions to create the Mg ATP complex, which binds to Fe protein and energises it to transfer electrons from the iron atoms in Fe protein to MoFe protein, prior to reaching the bound substrate.
In order to counterbalance this electron, an H+ ion generated by the dissociation of H2O becomes attached to the Mo atom of the MoFe protein. Repeatedly, electrons travel from flavodoxin to MoFe protein via Fe protein. Every time an electron is transported, two molecules of ATP are used.
Again, the second electron is balanced by an additional H+ ion. When the third electron is transferred to the MoFe protein, the H+ ions are displaced by nitrogen, resulting in the release of one hydrogen molecule. This third electron is once more neutralised by the attachment of one H+ ion to nitrogen, i.e. HN = NH.
This process of electron transfer is repeated until a total of eight electrons are transferred, thus reducing N to NH3. Figure depicts the sequence of reactions involving electron transport.
H+ ———— Counter balance 1st e– (electron)
H+ ————- Counter balance 2nd e– (electron)
H+ + H+ → H2 evolved
H+ …. N=N ….. H+ ← N=N
(3rd e–) (4th e–)
H+ … HN.—— .NH . H+
(5th e–) (6th e–)
H+ ….. H2N.———- .NH2 …….. H+
(7th e–) (8th e–)
NH3 + NH3
i.e. 2NH3
Thus, net 8 e– and 8H+ are required to reduce N2 to two ammonia molecules. Two moles of ATP are used per electron. Thus, 16 moles of ATP are utilised. The H2 evolution is mediated by a net transfer of two electrons, resulting in the depletion of four ATP molecules. Clearly, the nitrogen reduction process consumes 12 ATP, whereas the biological nitrogen fixation process is far more expensive.
Formally, the enzyme reaction should be written as follows:
N2 + 8H+ + 16 ATP + 8 e– → 2 NH3 + H2 + 16 ADP
Nitrogen fixation is a reductive process in which N2 is converted to the inorganic product NH3. These studies were validated by autoradiography (use of 15N2). On the other hand, glutamic acid is the first organic product generated.
Based on the oxidation number, the following scale is proposed:
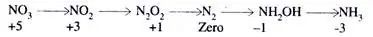
NO3 is highly oxidised on the above scale, while ammonia is significantly reduced. Therefore, a continuous reduction runs from oxidation number zero to oxidation number minus three during nitrogen fixation.
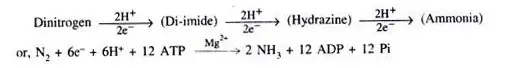
5. Hydrogen Evolution
One molecule of H2 is produced for every molecule of N2 that is fixed. This technique is costly for nitrogen fixation since it requires 16 ATP molecules, whereas a similar fixation can be accomplished with 12 ATP molecules.
It indicates that four molecules are being wasted. Hydrogenase, the enzyme responsible for regular hydrogen evolution (i.e., that created in the normal metabolism of these bacteria, not via nitrogenase) can catalyse both the uptake and evolution of hydrogen to circumvent this process.
The hydrogen is captured and recycled by hydrogenase in Azotobacter. The use of recycled hydrogen to produce additional ATP may contribute to Azotobacter’s highly efficient ATP economy.
In addition to cyanobacteria and photosynthetic bacteria, several rhizobia also exhibit photo-evolution of hydrogen. “Tight” symbiosis, which does not evolve H2, fixes more nitrogen per unit of solar energy than “loose” (H2-evolving) symbiosis. The non-leguminous relationships have been “tighter” than the majority of leguminous relationships.
References
- Vitousek, P. M., Menge, D. N. L., Reed, S. C., & Cleveland, C. C. (2013). Biological nitrogen fixation: rates, patterns and ecological controls in terrestrial ecosystems. Philosophical Transactions of the Royal Society B: Biological Sciences, 368(1621), 20130119–20130119. doi:10.1098/rstb.2013.0119
- https://www.nature.com/scitable/knowledge/library/biological-nitrogen-fixation-23570419/
- http://archive.bio.ed.ac.uk/jdeacon/microbes/nitrogen.htm
- https://eos.com/blog/nitrogen-fixation/
- https://www.cs.mcgill.ca/~rwest/wikispeedia/wpcd/wp/n/Nitrogen_fixation.htm
- https://www.biologydiscussion.com/nitrogen-fixation/types-nitrogen-fixation/nitrogen-fixation-types-physical-and-biological-nitrogen-fixation-with-diagram/14969
- https://www.biologydiscussion.com/plant-physiology-2/nitrogen-metabolism/mechanism-of-biological-nitrogen-fixation/22810
- https://unacademy.com/content/neet-ug/study-material/biology/biological-nitrogen-fixation/
- https://biologyboom.com/biological-nitrogen-fixation/
- https://www.biology-pages.info/N/NitrogenFixation.html
- https://en.wikipedia.org/wiki/Nitrogen_fixation
- https://www.britannica.com/science/nitrogen-fixation