Next-Generation Sequencing (NGS) is a technology for high-throughput DNA and RNA sequencing. It allows for the rapid and simultaneous analysis of millions of DNA fragments, enabling comprehensive genomic studies such as genome sequencing, transcriptome analysis, and epigenetic profiling. NGS has revolutionized the field of genomics, providing a faster and more cost-effective way to study the genetic information of organisms and facilitating the discovery of new biological insights.
Next Generation Sequencing (NGS) technology has changed the game in genetics research and scientific studies. With its ability to assess multiple genes in a single assay, NGS is now the go-to tool for clinical researchers and scientists. This technology has the power to sequence entire or targeted genomes in record time, making it a highly valuable resource for the scientific community.
But what exactly is NGS, and how does it work? In this article, we’ll dive into the details of this innovative technology and explore the ways it’s transforming the field of genetics.
NGS is a revolutionary tool that enables the rapid sequencing of large amounts of DNA. It works by breaking down the DNA into millions of small fragments, which are then sequenced in parallel using various sequencing technologies. The resulting data is then pieced together using bioinformatics analyses to produce a complete picture of the genome being studied.
The different NGS platforms use different sequencing technologies, but the basic principle of parallel sequencing remains the same. This makes NGS a versatile tool that can be used for a wide range of applications, from whole genome sequencing to targeted sequencing of specific genes of interest.
One of the key benefits of NGS is its speed and efficiency. By sequencing millions of fragments in parallel, NGS can produce results in a fraction of the time it would take to sequence a genome using traditional methods. This makes it an ideal tool for large-scale genomic studies, where time is of the essence.
Another major benefit of NGS is its ability to generate high-quality data. By sequencing multiple copies of each fragment, NGS reduces the chance of errors and provides a more accurate representation of the genome being studied. This is particularly important in applications such as diagnosing genetic diseases, where accurate information is critical.
Finally, NGS is highly cost-effective. The parallel sequencing approach used by NGS allows for the efficient use of resources, making it a more affordable option for many scientific studies. This has opened the doors for smaller research groups and institutions to participate in large-scale genomic studies, increasing the overall diversity of the scientific community.
Principle of Next-Generation Sequencing
Similar to Capillary Electrophoresis (CE) sequencing, Next Generation Sequencing technique involves the integration of fluorescently tagged deoxyribonucleotide triphosphates (dNTPs) into a DNA template strand during sequential cycles of DNA synthesis. Fluorophore excitation at the addition of each nucleotide is used to identify the nucleotides during each procedure. Instead of sequencing a single DNA fragment, next-generation sequencing simultaneously sequences millions of fragments. NGS provides excellent precision, a high rate of error-free readings, and a high proportion of base calls exceeding Q30.
Basic Steps Involved in Next-Generation Sequencing
Sequencing using an Illumina device involves four fundamental steps: Library Preparation, Cluster Generation, Sequencing, and Analysis. After the isolation of DNA or cDNA (synthesised from RNA), these four fundamental processes must be carried out:

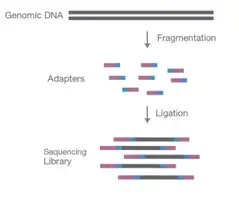
Preparing the samples to be compatible with the sequencer is a crucial step in the sequencing procedure, making library preparation a crucial step. Using either sonication or enzymatic restriction, DNA or cDNA samples are fragmented into 200-500 bp-long pieces. During fragmentation, each fragment acquires an A-tail overhang, which prepares them for ligation to the adapter sequence, which contains a T-base overhang complementary to the A-tail fragment.
Adapters are the sequence including primer binding sites, index sequences, and the sequence that enables library fragments to adhere to the flow cell lawn. These adapters have 5′ and 3′ ends that are ligated. Tagmentation combines fragmentation and ligation reactions in a single step, boosting the efficiency of the library preparation procedure.
During the PCR process, it aids in the enrichment of adapter-ligated DNA.
Individual adapters are used for each sample, and all samples are combined in a single tube. After pooling, the information is fed into the sequencer for further sequencing.
Cluster Generation
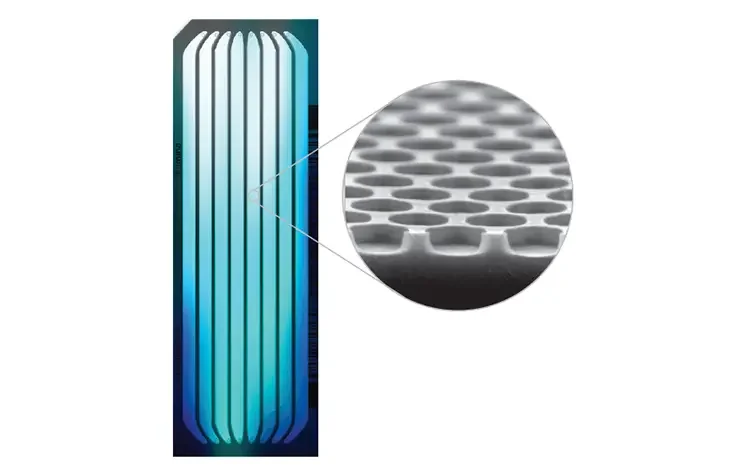
- The library is put into a flow cell and inserted into the sequencer. A flow cell is a glass slide with one, two, or eight physically separated lanes coated with an adapter-complementary oligo lawn.
- Currently, the most common type of flow cell is a patterned flow cell manufactured with semiconductor fabrication techniques. It has a glass substrate with patterned nano wells holding DNA probes that grab the prepared DNA strands during cluster creation for amplification.
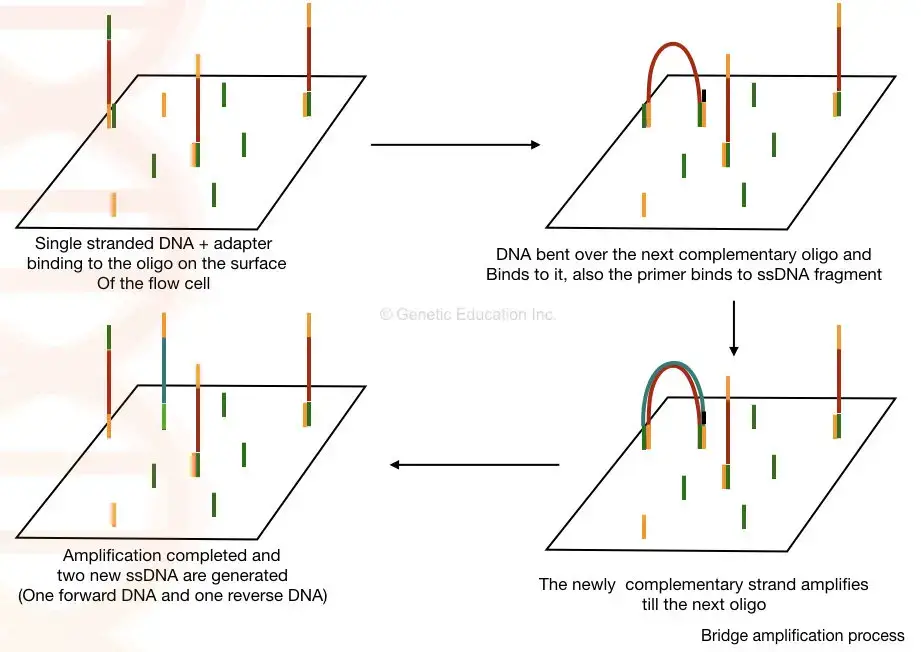
- Once samples are coupled to the flow cell and bridge amplification occurs, cluster formation commences. During this procedure, DNA fragments from the library hybridise with an oligonucleotide on the flow cell surface.
- DNA polymerase is subsequently used to generate a complementary strand by elongating the oligo linked to the flow cell. The initial molecule is washed away, and the strand connects to the next oligo in the flow cell by bending over like a bridge.
- This second oligo is complementary to another adapter sequence, and polymerase forms a double-stranded bridge by generating a complimentary strand. This bridge is denatured, resulting in two single-stranded copies of the attached molecule. This procedure is performed repeatedly and concurrently for millions of clusters, resulting in the clonal amplification of all.
- The reverse strands are severed and removed after bridge amplification, leaving only the forward strands. To prevent unintended priming, the 3′ ends are blocked. After completion of cluster creation, the templates are ready for sequencing.
Sequencing by Synthesis
- Each cluster on a flow cell has a matching sequencing. Sequencing requires sequencing primer, DNA polymerase, and a fluorophore that has been fluorescently tagged.
- Depending on the chemistry utilised in their different machines (4-channel chemistry, 2-channel chemistry, and 1-channel chemistry), either all nucleotides are labelled with the fluorophore or only a few nucleotides are labelled.
- All nucleotides are tagged with four fluorescent dyes in 4-channel chemistry. Two-channel chemistry employs two fluorescent dyes, whereas one-channel chemistry employs only one.
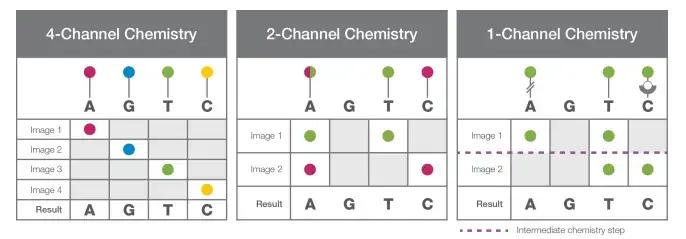
- In 4-channel chemistry, all the aforementioned components are processed through the flow cell, where sequencing primer anneals to its corresponding position on the adaptor and DNA polymerase adds the fluorescently-labeled complementary nucleotides.
- This fluorophore works as a blocking group, preventing DNA polymerase from adding another nucleotide until it is removed, so enabling the detector to record the fluorescence of each base being added.
- After each nucleotide is added, the light source excites the fluorophore in the cluster, and the characteristic fluorescence signal emitted is recorded. This method is known as sequencing-by-synthesis.
- Only one strand of DNA from this cluster is depicted in the illustration, however the sequencing occurs simultaneously in the complete cluster. DNA polymerase, fluorescently tagged dNTPs, and sequencing primers are added as illustrated.
- DNA polymerase attaches primer to the DNA template, which is then stretched. Once the complementary nucleotide has been added to the end of the primer, fluorescence prevents DNA polymerase from adding additional nucleotides, and the computer records the fluorophore.
- This fluorescence is then eliminated by washing. DNA polymerase then incorporates the second complementary nucleotide; these cycles are repeated in order to obtain a read from this cluster. DNA Sequencing Unit. Molecular Biology is the source.
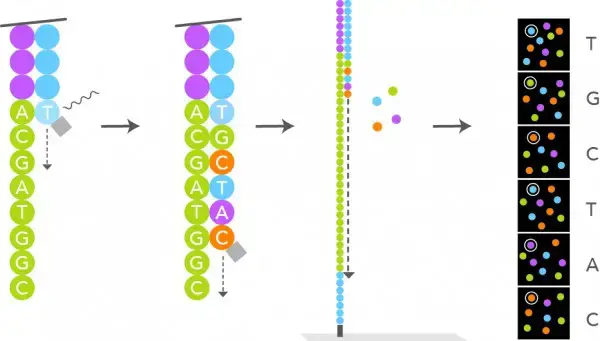
How to read the nucleotide in NGS?
- Different lengths of DNA fragments are read at various stages during sequencing, and each cluster has numerous reads.
- Once the flow cell is flooded with all of the components (DNA polymerase, sequencing primer, and fluorescently tagged nucleotide), the sequencing primer anneals to the left side of genomic DNA, which reads the fragment from the 5′ side and is regarded as the first read or read 1. After removing these components, the 3′ end of the template is deprotected, allowing it to fold over and bind to the oligo on a flow cell.
- The flow cell is once again inundated with all materials, but primers corresponding to the appropriate adaptor are added. This constitutes a second read or read 2 as it decodes the index discovered in the compatible adaptor.
- After each cycle, an optical device detects fluorophore emission from each cluster to determine the base incorporated. This method of finding the sequence from both ends is known as paired-end sequencing.
- In contrast, single-read sequencing determines the sequences from only one end of a library fragment.
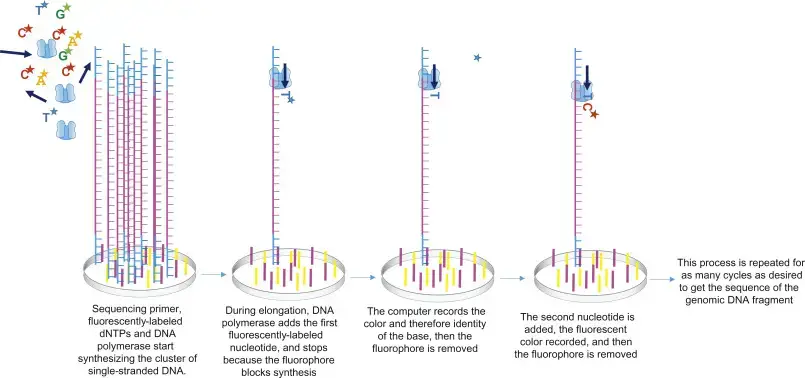
Data Analysis
- Following the completion of sequencing, the optical signals are translated into a nucleotide sequence, a process known as base calling. The Phred quality score (Q score), the most prevalent metric for evaluating the quality of sequencing data, is used to quantify the precision of base calling. The Q score represents the likelihood that a specific base is mistakenly named by the sequencer.
- Q scores are logarithmically related to the base calling error probability (P)2. Q= -10 log10P
- This Q score defines if a base is good or awful. Below is displayed the Quality Score and Base Calling Accuracy. The Q30 is ideally suited for a variety of sequencing applications.
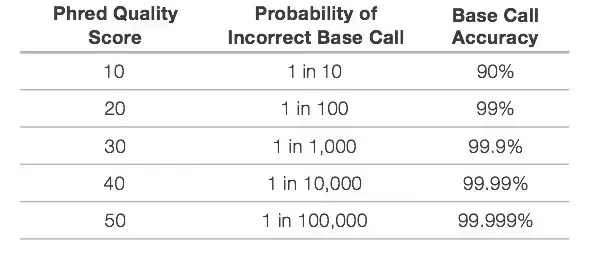
- During library preparation, unique index sequences are assigned to each sample; this is known as multiplexing, and it enables a large number of libraries to be pooled together and sequenced simultaneously in a single sequencing run.
- Therefore, prior to data analysis, demultiplexing happens, which separates sequences from sample libraries based on their unique indices. Reads with comparable sequences of bases are grouped locally for each sample.
- Together, forward and reverse reads create continuous sequences. These recently discovered contiguous sequences are aligned with a reference sequence.
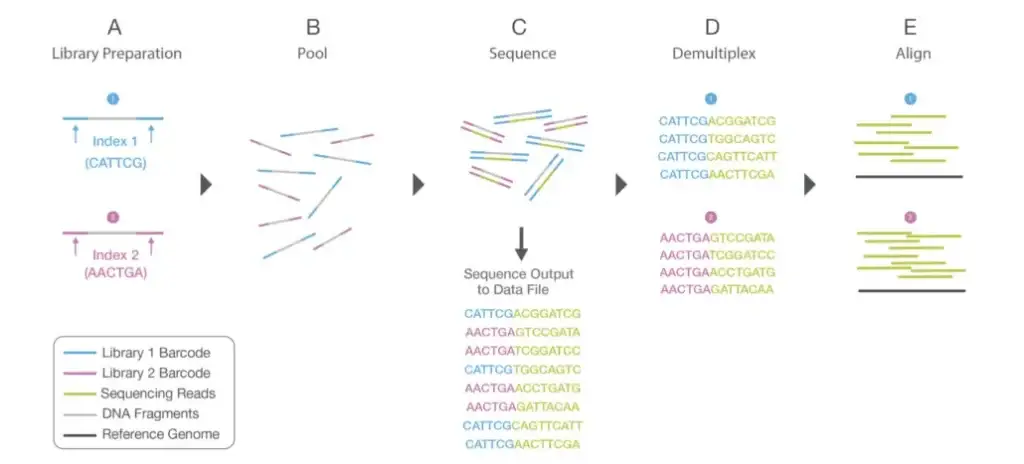
- Important is the number of reads that align with the reference genome, or coverage depth. The greater the coverage depth, the greater the number of sequences that are identical and aligned. This alignment illustrates bioinformatics-identifiable differences and similarities between the reference genome and the sequencing of isolated samples.
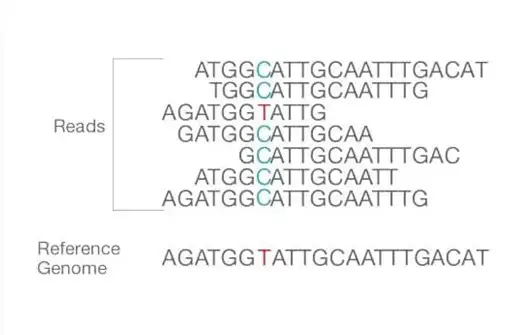
- After alignment, numerous analysis variations are conceivable, including identification of single nucleotide polymorphism (SNP) or insertion-deletion (indel), read counting for RNA approaches, phylogenetic or metagenomic analysis, and others.
Application of Next-Generation Sequencing
The technology of Next Generation Sequencing has numerous uses. Several of these are listed below:
- NGS can be used to do metagenomic sequencing (the technique of identifying organisms from environmental or clinical samples by utilising numerous sets of primers for various species) for the detection of undiscovered disease-associated viruses and novel human viruses.
- In humans, less than 2% of the genome comprises an exome containing the majority of known disease-causing mutations, and entire exome sequencing using NGS is cost-effective.
- This approach is used to sequence the genomes of non-human animals, such as agriculturally significant livestock, plants, and microorganisms associated with illness.
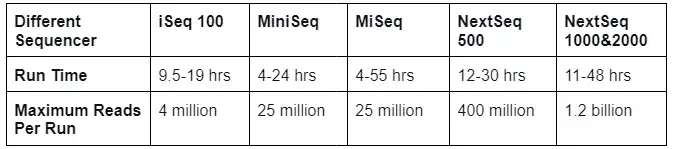
- Whole genome sequencing is a significant tool for genomics research since it analyses the entire genome and generates vast amounts of data. Different sequencers exist that can sequence a sample in a short amount of time, as listed below:
Advantages of Next-Generation Sequencing
Next-Generation Sequencing (NGS) has several advantages over traditional sequencing methods, including:
- High throughput: NGS allows for the simultaneous analysis of millions of DNA sequences, leading to faster and more comprehensive data generation.
- Increased accuracy: NGS technology allows for the detection of even low-frequency genetic variations.
- Reduced cost: The cost of NGS has decreased dramatically in recent years, making it more accessible and affordable for a wider range of applications.
- Improved versatility: NGS can be applied to a variety of sample types, including whole genomes, exomes, transcriptomes, and epigenomes.
- Scalability: NGS can be scaled up or down depending on the needs of the experiment, making it suitable for large-scale population studies or small, targeted analyses.
- Improved data analysis: Advanced computational methods have been developed to analyze NGS data, leading to improved understanding of biological systems.
Limitation of Next-Generation Sequencing
NGS has several restrictions, which are outlined below:
- As it involves complex bioinformatics tools, rapid data processing, and enormous data storage capacities, it can be expensive.
- PCR amplification prior to sequencing might result in PCR biases during library preparation (GC-content, fragment length, and false diversity) and analysis (base errors/favoring particular sequences over others).
FAQ
What is Next-Generation Sequencing (NGS)?
Next-Generation Sequencing (NGS) is a high-throughput DNA and RNA sequencing technology that enables the rapid and simultaneous analysis of millions of DNA fragments, allowing for comprehensive genomic studies.
How does NGS work?
NGS works by breaking down DNA into small fragments, which are then sequenced in parallel using various sequencing technologies. The resulting data is pieced together using bioinformatics to produce a complete picture of the genome being studied.
What are the benefits of NGS?
The benefits of NGS include speed and efficiency, the generation of high-quality data, and cost-effectiveness.
What is the principle of Next-Generation Sequencing?
The principle of Next-Generation Sequencing involves integrating fluorescently tagged deoxyribonucleotide triphosphates into a DNA template strand during sequential cycles of DNA synthesis. Millions of fragments are simultaneously sequenced.
What are the basic steps involved in Next-Generation Sequencing?
The basic steps involved in Next-Generation Sequencing include Library Preparation, Cluster Generation, Sequencing, and Analysis. Library preparation involves preparing samples to be compatible with the sequencer, followed by adapter ligation and PCR enrichment.
References
- Clark D, Pazdernik N, McGehee M. DNA Sequencing Unit. Molecular Biology. 2019. 240–269 p.
- https://www.illumina.com/science/technology/next-generation-sequencing/beginners/ngs-workflow.html
- https://medium.com/@tiffanysouterre/dna-sequencing-techniques-explained-53c21eef51b1
- https://www.cd-genomics.com/blog/principle-and-workflow-of-illumina-next-generation-sequencing/
- Illumina sites
- https://www.technologynetworks.com/genomics/articles/an-overview-of-next-generation-sequencing