What is Lineweaver–Burk plot?
The Lineweaver–Burk plot, commonly referred to as the double reciprocal plot, stands as a foundational concept within the domain of biochemistry. Stemming from the renowned Michaelis–Menten equation, which pertains to enzyme kinetics, this plot was innovatively introduced by Hans Lineweaver and Dean Burk in the year 1934. Delving into its essence, the Lineweaver–Burk plot is fundamentally a graphical portrayal of the Michaelis–Menten equation. Therefore, its understanding is pivotal for those deeply invested in comprehending the intricacies of enzyme kinetics.
However, a noteworthy observation that emerges from empirical findings is the distortion of the error structure of the data within this double reciprocal plot. Therefore, although the Lineweaver–Burk plot has earned its historical significance for the evaluation of enzymatic parameters, its accuracy has come under scrutiny. Besides this, even other linear forms of the Michaelis–Menten equation, such as the Hanes–Woolf plot and the Eadie–Hofstee plot, have their limitations. Hence, researchers have advised caution when employing all linearized forms of the Michaelis–Menten equation for the calculation of kinetic parameters. Then, with the dawn of the digital era and the consequent ubiquitous availability of desktop computers, non-linear regression methods, which are weighted appropriately, have risen to prominence. These methods, by virtue of their enhanced accuracy, have been largely preferred over traditional approaches.
To elucidate the functional mechanism behind the Lineweaver–Burk plot, one must first grasp the Michaelis–Menten equation, represented as:
\( v = \frac{V_a}{K_m + a} \)
In this equation, the rate v is determined by the substrate concentration a and two pivotal parameters: V, which represents the limiting rate, and Km, symbolizing the Michaelis constant. An intriguing transformation of this equation is achieved by taking the reciprocals of both its sides. This metamorphosis leads to the expression:
\( \frac{1}{v} = \frac{1}{V} + K_m \cdot \frac{1}{a} \)
Therefore, the Lineweaver–Burk plot is created by plotting 1/v against 1/a. The resultant graphical representation is a straight line characterized by an ordinate intercept of 1/V, an abscissa intercept of -1/Km, and a slope which is determined by the ratio Km/V.
In summary, the Lineweaver–Burk plot, while historically revered, has certain limitations in terms of accuracy. Nevertheless, its conceptual significance in elucidating enzyme kinetics remains undiminished. The evolution of computational tools has facilitated more precise methodologies, emphasizing the dynamic and evolving nature of scientific understanding.
Lineweaver–Burk Plot
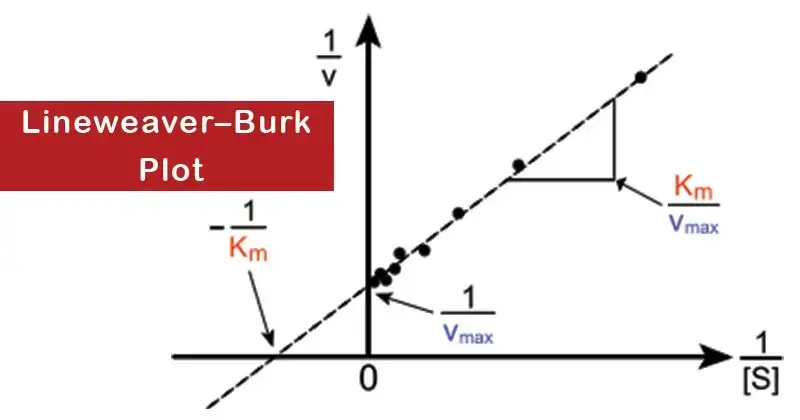
The study of enzyme kinetics has always been a crucial component of biochemistry. Within this realm, the accurate determination of Vmax and Km from a hyperbolic plot poses a challenge, particularly given that Vmax is realized at infinite substrate concentration. Therefore, to circumvent this complexity, Lineweaver and Burk proposed a transformation of the Michaelis–Menten equation, resulting in a straight-line equation. This transformative approach culminated in what is now widely recognized as the Lineweaver–Burk plot, or the double reciprocal plot, first described in 1934 by Hans Lineweaver and Dean Burk.
Diving into the technicalities, the Lineweaver–Burk equation is rooted in the Michaelis–Menten equation and can be presented as:
\( \frac{1}{V} = \frac{K_m + [S]}{V_{\text{max}} [S]} = \frac{K_m}{V_{\text{max}} [S]} + \frac{1}{V_{\text{max}}} \)
In this equation, V symbolizes the reaction velocity or rate, while Km represents the Michaelis–Menten constant. Vmax denotes the maximum reaction velocity, and [S] stands for the substrate concentration. When plotted, this equation yields a straight line. The y-axis intercept corresponds to 1/Vmax1 and the x-axis intercept equates to −1/Km−1. The slope of this straight line is given by Km/Vmax.
To generate a Lineweaver–Burk plot, one commences by measuring V0 at varying substrate concentrations. Subsequently, a plot of 1/V0 against 1/[S] is crafted.
Moreover, the Lineweaver–Burk plot serves another vital function: discerning the nature of enzyme inhibitors, whether they are competitive or noncompetitive. This distinction is paramount because it illuminates the manner in which an inhibitor associates with an enzyme. By analyzing V0 at multiple substrate concentrations, especially in the presence of a constant inhibitor concentration, one can decipher the type of inhibition:
- Competitive Inhibition: Within the Lineweaver–Burk plot, competitive inhibitors manifest by increasing the slope of the line. Additionally, the x-axis intercept undergoes alteration due to an increase in Km, while the y-axis intercept remains steadfast, signifying that Vmax remains unaltered.
- Noncompetitive Inhibition: On the other hand, noncompetitive inhibitors influence the plot differently. They increase the plot’s slope and modify the y-axis intercept, indicating a decrease in Vmax. Contrarily, the x-axis intercept remains consistent, denoting that Km remains unchanged.
In essence, the Lineweaver–Burk plot stands as an invaluable tool, not just for understanding the fundamental parameters of enzyme kinetics but also for distinguishing between different types of enzyme inhibitors.
Applications of Lineweaver–Burk Plot
In the realm of biochemistry, the Lineweaver–Burk plot stands as an instrumental tool, offering a nuanced approach to understanding enzyme kinetics. This graphical representation has a plethora of applications which can be categorized and described in detail as follows:
- Determination of Kinetic Parameters: Before the advent of advanced computers and sophisticated non-linear regression software, the Lineweaver–Burk plot was the premier choice for ascertaining vital terms in enzyme kinetics, particularly Km and Vmax. Therefore, this plot allowed scientists to visualize and quantify the behavior of enzymes with respect to substrates in a systematic and logical manner.
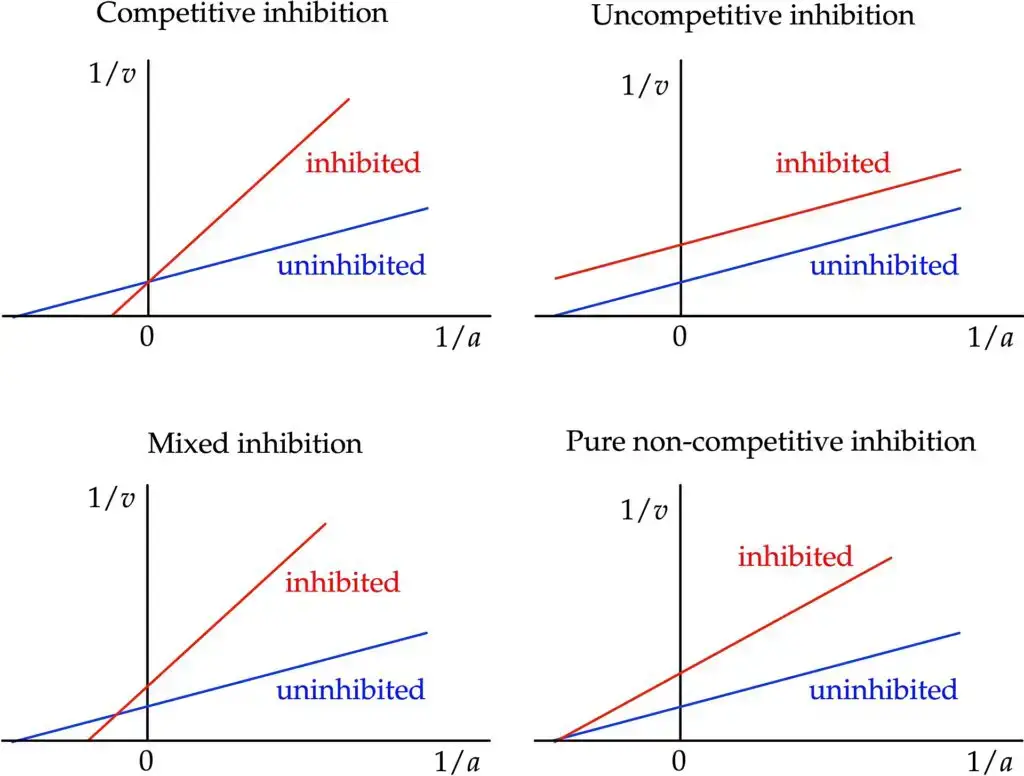
- Visual Insight into Enzyme Inhibition: Besides its use in identifying key kinetic parameters, the Lineweaver–Burk plot offers a swift, visual impression of the diverse forms of enzyme inhibition. When comparing various modes of inhibition, this plot becomes indispensable, providing a clear contrast to the uninhibited reaction.
- Competitive Inhibition: Within this type of inhibition, the apparent value of Vmax remains unchanged, leading to competitive inhibitors sharing the same intercept on the ordinate as uninhibited enzymes. Consequently, competitive inhibition amplifies the apparent Km value, denoting reduced substrate affinity. Graphically, this translates to the inhibited enzyme having an expanded intercept on the abscissa in the plot.
- Pure Non-Competitive Inhibition: This form of inhibition sees a reduction in the apparent value of Vmax. On the Lineweaver–Burk plot, this manifests as an augmented ordinate intercept. However, it leaves the abscissa intercept, −1/Km, unaffected, emphasizing that substrate affinity is unaltered in this scenario.
- Mixed Inhibition: In stark contrast to pure non-competitive inhibition, mixed inhibition affects both Vmax and Km. Typically, Km experiences an increase, implying a decline in substrate affinity with mixed inhibition. It’s worth noting that while pure noncompetitive inhibition exists, it’s relatively rare. Hence, Cleland redefined noncompetitive to mean mixed, a terminology adopted by many, but not universally.
- Uncompetitive Inhibition: Here, there’s a noticeable decline in the apparent Vmax value, and the ratio V/Km is also influenced. On the Lineweaver–Burk plot, this appears as an increased ordinate intercept without any change in the slope. Furthermore, substrate affinity heightens with uncompetitive inhibition, reducing the apparent Km value. A hallmark of uncompetitive inhibition in the plot is the emergence of parallel lines for varying inhibitor concentrations.
References
- https://www.medschoolcoach.com/lineweaver-burk-plots-mcat-biochemistry/
- https://en.wikipedia.org/wiki/LineweaverBurk_plot
- https://www.degruyter.com/database/IUPAC/entry/iupac.90.0327/html
- https://www.bionity.com/en/encyclopedia/Lineweaver-Burk_plot.html