Gram-Sure Test is a rapid qualitative diagnostic test used for the differentiation of aerobic Gram-negative and Gram-positive bacteria. It is mainly applied for bacteria that are rods or coccobacilli in shape. This test works by detecting the presence of a specific enzyme known as L-alanine aminopeptidase. This enzyme is present almost exclusively in Gram-negative bacterial cell wall and generally absent in Gram-positive bacteria.
It is the process in which reagent impregnated disks containing a fluorogenic substrate, L-alanine-7-amido-4-methylcoumarin, is used. When the bacterial sample is applied on the disk, the enzyme is released by Gram-negative organisms. The enzyme hydrolyzes the substrate and forms 7-amino-4-methylcoumarin. This compound produces a bright blue fluorescence when observed under long wave ultraviolet light (360 nm). The reaction is usually completed within 5–10 minutes at room temperature.
This test is particularly useful as a confirmatory method for organisms showing Gram-variable staining reaction. These are bacteria which are difficult to identify clearly by Gram staining such as Bacillus, Listeria, and Erysipelothrix species. However, this test is not suitable for obligate anaerobic bacteria like Bacteroides and Fusobacterium as they do not give reliable results in this method.
Objectives of Gram-Sure Test
- To differentiate between aerobic Gram-negative and Gram-positive bacteria, mainly rods or coccobacilli.
- To detect the presence of L-alanine aminopeptidase enzyme present in the cell wall of Gram-negative bacteria.
- To identify bacteria which show Gram-variable staining reaction or give unclear Gram stain results.
- To provide a rapid confirmatory result within few minutes as compared to other biochemical methods.
- To support and confirm the results of traditional Gram staining method and not to replace it.
Principle of Gram-Sure
It is based on the detection of the enzyme L-alanine aminopeptidase which is localized in the cell wall of Gram-negative bacteria. This enzyme is generally absent or remains inactive in Gram-positive organisms. The presence or absence of this enzyme forms the basic principle of Gram-Sure test.
In this test, disks impregnated with L-Alanine-7-amido-4-methylcoumarin are used. This substrate is non-fluorescent in nature. When Gram-negative bacteria are present, the aminopeptidase enzyme hydrolyzes the substrate by cleaving L-alanine from it. This reaction releases 7-amino-4-methylcoumarin.
The released compound produces a bright blue fluorescence when exposed to long-wave ultraviolet light. The appearance of fluorescence indicates a positive reaction showing Gram-negative bacteria. Absence of fluorescence indicates a negative reaction and confirms Gram-positive bacteria.
Materials Required
- Gram-Sure disks impregnated with L-alanine-7-amido-4-methylcoumarin.
- Demineralized or deionized water (about 0.25 ml per test).
- Fresh pure culture of test organism (preferably less than 48 hours old).
- Quality control organisms – Escherichia coli (positive control) and Staphylococcus aureus (negative control).
- Small test tubes (10 × 75 mm size).
- Inoculating loop or sterile swab.
- Standard containers for collection of samples.
- Long-wave ultraviolet light source (around 360 nm) for observing fluorescence.
- Loop sterilization device to maintain aseptic condition.
- Incubator or laboratory setup for room temperature incubation.
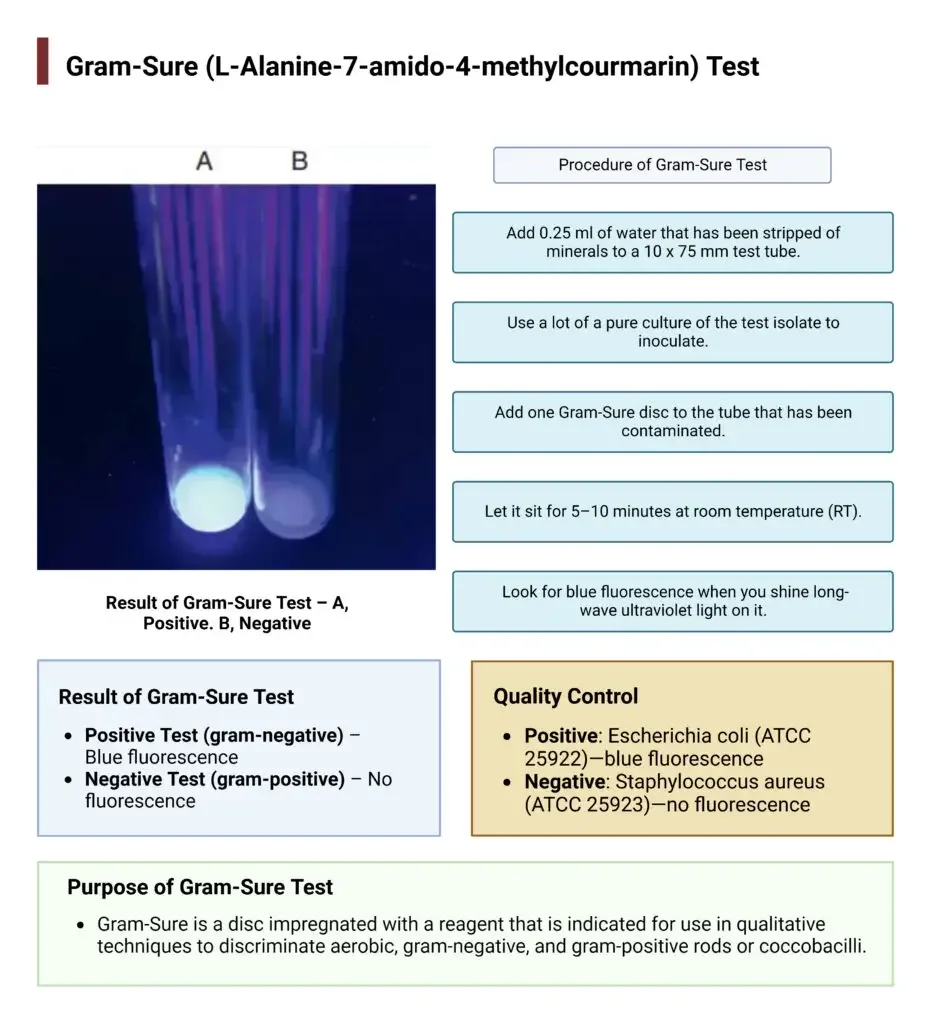
Procedure of Gram-Sure Test
- Take a clean test tube (10 × 75 mm) and add 0.25 ml of demineralized or deionized water.
- Pick a colony from a fresh pure culture with sterile inoculating loop or swab.
- Mix the culture in water to make a heavy milky suspension.
- Place one Gram-Sure reagent disk into the suspension aseptically.
- Allow the tube to stand at room temperature for 5–10 minutes.
- Observe the tube under long-wave ultraviolet light (360 nm).
- Blue fluorescence indicates Gram-negative bacteria.
- No fluorescence indicates Gram-positive bacteria.
Result of Gram-Sure Test
Positive Result (Gram-Negative)
- Observation – A bright blue fluorescence is seen when the test tube is viewed under long-wave UV light (about 360 nm).
- It is the indication that the enzyme L-alanine aminopeptidase is present in the organism.
- These are aerobic Gram-negative rods or coccobacillus (Escherichia coli, Pseudomonas aeruginosa).
Negative Result (Gram-Positive)
- Observation – No fluorescence is produced and the suspension remain unchanged under UV light.
- It is the process where the organism does not have L-alanine aminopeptidase activity.
- These organism is Gram-positive (Staphylococcus aureus, Bacillus species).
Exceptions and Limitations
- Obligate anaerobes – These organisms like Bacteroides, Fusobacterium, and Veillonella does not produce predictable reactions, so it is not tested by this method.
- Campylobacter species – This is Gram-negative but the enzyme is absent, so the test result is negative.

Organisms showing result in Gram-Sure Test
Positive result
(It indicates Gram-negative bacteria due to presence of L-alanine aminopeptidase enzyme)
- Escherichia coli (Quality control strain).
- Pseudomonas aeruginosa (Quality control strain).
- Klebsiella pneumoniae.
- Proteus mirabilis.
- Most aerobic and facultatively anaerobic Gram-negative rods and coccobacilli.
Negative result
(It indicates Gram-positive bacteria due to absence of L-alanine aminopeptidase enzyme)
- Staphylococcus aureus (Quality control strain).
- Enterococcus faecalis (Quality control strain).
- Bacillus species (for example Bacillus subtilis).
- Listeria species (for example Listeria monocytogenes).
- Lactobacillus species.
- Erysipelothrix species.
- Most Gram-positive rods and coccobacilli including Gram-variable organisms.
Exceptions
(Gram-negative bacteria showing negative result due to lack of enzyme or unpredictable reaction)
- Campylobacter species.
- Bacteroides species (for example Bacteroides fragilis and Bacteroides vulgatus).
- Veillonella parvula.
- Helicobacter species.
- Arcobacter species.
- Obligate anaerobic organisms (generally unsuitable for this test).
Quality Control
Positive control (Gram-negative organism)
- Escherichia coli ATCC® 25922.
- Pseudomonas aeruginosa ATCC® 27853 (alternative strain).
- Expected result is bright blue fluorescence under long wave ultraviolet light (360 nm).
Negative control (Gram-positive organism)
- Staphylococcus aureus ATCC® 25923.
- Enterococcus faecalis ATCC® 29212 (alternative strain).
- Expected result is absence of fluorescence.
Uses of Gram-Sure Test
- It is used for qualitative differentiation of aerobic Gram-negative and Gram-positive rods or coccobacilli.
- It is helpful in clarification of doubtful or Gram-variable staining reaction where Gram stain result is unclear.
- It is used to prevent misclassification of Gram-positive rods which appear Gram-negative or Gram-variable such as Bacillus Erysipelothrix Lactobacillus and Listeria.
- It is used as a confirmatory or supplementary test along with conventional Gram staining method and not as a replacement.
- It is used for rapid screening of Gram reaction giving result within 5 to 10 minutes as compared to antibiotic susceptibility based methods.
Advantages of Gram-Sure Test
- It gives rapid result within 5 to 10 minutes at room temperature as compared to other confirmatory methods.
- It is easy to perform as ready-to-use reagent impregnated discs are provided and no special preparation is required.
- It shows high sensitivity due to use of fluorogenic substrate (L-alanine-7-amido-4-methylcoumarin).
- It gives clear and easy interpretation as Gram-negative organisms show bright blue fluorescence while Gram-positive organisms do not fluoresce.
- It is useful in resolving Gram-variable or poorly staining organisms such as Bacillus Erysipelothrix Lactobacillus and Listeria.
- It is a better predictor of Gram reaction as compared to vancomycin or colimycin susceptibility testing in difficult groups.
- It is cost effective and less expensive than antibiotic susceptibility based confirmatory methods.
Limitation of Gram-Sure Test
- It is not suitable for obligate anaerobic organisms such as Bacteroides Fusobacterium and Veillonella as these organisms lack required enzyme activity.
- It gives false negative result with some microaerophilic Gram-negative bacteria like Campylobacter and Helicobacter species.
- It is not recommended for Gram-positive cocci such as Streptococcus as results may be unpredictable or false positive.
- Colonies grown on media containing dyes or indicators like MacConkey agar or EMB agar should not be used as dyes may interfere with fluorescence.
- It does not provide information about bacterial morphology or arrangement unlike conventional Gram staining.
- It requires long wave ultraviolet light source (around 360 nm) for observation of fluorescence.
- It is not a definitive identification test and further biochemical or serological tests are required for confirmation.
- Beveridge, T. J. (1999). Structures of Gram-negative cell walls and their derived membrane vesicles. Journal of Bacteriology, 181(16), 4725–4733. https://doi.org/10.1128/jb.181.16.4725-4733.1999
- BOC Sciences. (n.d.). Coumarin-based fluorescent probes for imaging. https://www.bocsci.com/resources/coumarin-based-fluorescent-probes.html
- Breidenbach, J., Bartz, U., & Gütschow, M. (2020). Coumarin as a structural component of substrates and probes for serine and cysteine proteases. Biochimica et Biophysica Acta (BBA) – Proteins and Proteomics, 1868(9), 140445. https://doi.org/10.1016/j.bbapap.2020.140445
- Cava, F., Lam, H., de Pedro, M. A., & Waldor, M. K. (2010). Emerging knowledge of regulatory roles of d-amino acids in bacteria. Cellular and Molecular Life Sciences, 68(5), 817–831. https://doi.org/10.1007/s00018-010-0571-8
- Chi, H. (2014). A study of synthesis and application of peptide 4-methylcoumaryl-7-amide as the novel fluorescent substrates [Doctoral dissertation, Kyushu Institute of Technology].
- Ciancaglini, E., Fazii, P., & Sforza, G. R. (2004). The use of a differential fluorescent staining method to detect bacteriuria. Clinical Laboratory, 50(11-12), 685–688.
- Cornell University College of Veterinary Medicine. (2023, December). Gram stain protocol. https://www.vet.cornell.edu/animal-health-diagnostic-center/testing/testing-protocols-interpretations/gram-stain-protocol
- Creative Enzymes. (n.d.). L-Alanine 7-amido-4-methylcoumarin, trifluoroacetate salt. https://www.creative-enzymes.com/product/lalanine-7amido4methylcoumarin-trifluoroacetate-salt_8690.html
- Dik, D. A., Fisher, J. F., & Mobashery, S. (2018). Cell-wall recycling of the Gram-negative bacteria and the nexus to antibiotic resistance. Chemical Reviews, 118(12), 5952–5984. https://doi.org/10.1021/acs.chemrev.8b00277
- Drag, M., Bogyo, M., Ellman, J. A., & Salvesen, G. S. (2010). Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. Journal of Biological Chemistry, 285(5), 3310–3318. https://doi.org/10.1074/jbc.M109.060418
- Gist-Brocades N.V. (1997). Gram-negative alkaliphilic microorganisms (European Patent No. EP0473217B1). European Patent Office.
- Gonzales, T., & Robert-Baudouy, J. (1996). Bacterial aminopeptidases: Properties and functions. FEMS Microbiology Reviews, 18(4), 319–344.
- Hardy Diagnostics. (n.d.). LanaGram™ instructions for use (IFU-10518[A]).
- Johnson, J. W., Fisher, J. F., & Mobashery, S. (2013). Bacterial cell-wall recycling. Annals of the New York Academy of Sciences, 1277(1), 54–75. https://doi.org/10.1111/j.1749-6632.2012.06813.x
- Leonard, R. B., Ohlson, S., Newcomb-Gayman, P. L., & Carroll, K. C. (1995, August). Comparison of a commercial disk test with vancomycin and colimycin susceptibility testing for identification of bacteria with abnormal Gram staining reactions [Abstract]. ResearchGate.
- LifeTein. (2025, December 11). Unusual amino acids: Propargylglycine (Pra). LifeTein Peptide Blog. https://lifetein.com/blog/category/peptide_synthesis/
- Mandela, E., Stubenrauch, C. J., Ryoo, D., Hwang, H., Cohen, E. J., Torres, V. L., Deo, P., Webb, C. T., Huang, C., Schittenhelm, R. B., Beeby, M., Gumbart, J. C., Lithgow, T., & Hay, I. D. (2022). Adaptation of the periplasm to maintain spatial constraints essential for cell envelope processes and cell viability. eLife, 11, e73516. https://doi.org/10.7554/eLife.73516
- Merck Millipore. (n.d.). Bactident® Aminopeptidase [Technical data sheet].
- Merck Millipore. (2023). Technical data sheet Bactident® Aminopeptidase for the detection of L-alanine aminopeptidase in microorganisms.
- Miller, S. I., & Salama, N. R. (2018). The Gram-negative bacterial periplasm: Size matters. PLOS Biology, 16(1), e2004935. https://doi.org/10.1371/journal.pbio.2004935
- Paschalidou, K., Neumann, U., Gerhartz, B., & Tzougraki, C. (2004). Highly sensitive intramolecularly quenched fluorogenic substrates for renin based on the combination of L-2-amino-3-(7-methoxy-4-coumaryl)propionic acid with 2,4-dinitrophenyl groups at various positions. Biochemical Journal, 382(Pt 3), 1031–1038. https://doi.org/10.1042/BJ20040729
- Richter, P., Merz, A., Biboy, J., Paczia, N., … & Thanbichler, M. (2025). Peptidoglycan recycling is critical for cell division, cell wall integrity and β-lactam resistance in Caulobacter crescentus. eLife. https://doi.org/10.7554/eLife.109465
- Sharma, S., Acharya, J., Banjara, M. R., Ghimire, P., & Singh, A. (2020). Comparison of acridine orange fluorescent microscopy and Gram stain light microscopy for the rapid detection of bacteria in cerebrospinal fluid. BMC Research Notes, 13, 29. https://doi.org/10.1186/s13104-020-4895-7
- Sigma-Aldrich. (n.d.). 75554 Aminopeptidase test (Gram-positive-test) [Product information].
- Sigma-Aldrich. (n.d.). Aminopeptidase test strips suitable for agriculture, clinical testing, environmental, food and beverages, pharmaceutical, microbiology and specific enzyme detection. https://www.sigmaaldrich.com/US/en/product/sial/75554
- Sigma-Aldrich. (n.d.). L-Alanine 7-amido-4-methylcoumarin trifluoroacetate salt [Product information].
- Starzak, K., Matwijczuk, A., Creaven, B., Matwijczuk, A., Wybraniec, S., & Karcz, D. (2019). Fluorescence quenching-based mechanism for determination of hypochlorite by coumarin-derived sensors. International Journal of Molecular Sciences, 20(2), 281. https://doi.org/10.3390/ijms20020281
- Thermo Fisher Scientific. (2012). Oxoid and Remel catalog (Canada) 2012-2013.
- Thermo Fisher Scientific. (2021). Gram-Sure [Instructions for Use].
- Thermo Fisher Scientific. (n.d.). Thermo Scientific™ Gram-Sure. https://www.fishersci.com/shop/products/remel-gram-sure/R211820
- Torrens, G., & Cava, F. (2024). Mechanisms conferring bacterial cell wall variability and adaptivity. Biochemical Society Transactions, 52(5), 1981–1993. https://doi.org/10.1042/BST20230027
- Walsh, C. T. (1989). Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. Journal of Biological Chemistry, 264(5), 2393–2396.
- Webs UAB. (n.d.). Disks and strips for microbiology [Fluka Manual]. https://webs.uab.cat/workshopmrama/wp-content/uploads/sites/312/2011/06/disks_strips.pdf
- Wikipedia. (n.d.). Alanine aminopeptidase. Retrieved from https://en.wikipedia.org/wiki/Alanine_aminopeptidase
- Wu, H.-M., Cordeiro, S. M., Harcourt, B. H., Carvalho, M., Azevedo, J., Oliveira, T. Q., … & Mayer, L. W. (2013). Accuracy of real-time PCR, Gram stain and culture for Streptococcus pneumoniae, Neisseria meningitidis and Haemophilus influenzae meningitis diagnosis. BMC Infectious Diseases, 13, 26.
- Xu, X., Zhu, A., Bai, L., Li, S., Ge, W., Yang, Y., Liu, X., Qin, Z., Li, Z., & Li, J. (2025). Separation and detection of Gram-negative bacteria via vancomycin-functionalized magnetic beads and aminopeptidase test strips. Frontiers in Bioengineering and Biotechnology, 13, 1712799. https://doi.org/10.3389/fbioe.2025.1712799
- Zhang, J., Allen, J., Ward, S. J., Dekker, L. V., & Dreveny, I. (2025). A versatile fluorescence polarization-based deubiquitination assay using an isopeptide bond substrate mimetic (IsoMim). Journal of Biological Chemistry, 301(7), 110342. https://doi.org/10.1016/j.jbc.2025.110342
- Zhang, Y., Li, C., Jiang, M., Liu, Y., & Sun, Z. (2025). Advancements and prospects of metal-organic framework-based fluorescent sensors. Biosensors, 15(11), 709. https://doi.org/10.3390/bios15110709