What is gene expression?
- Gene expression is a fundamental biological process that dictates the functionality of cells and, by extension, the overall physiology of an organism. At its core, gene expression is the mechanism by which the genetic information encoded within DNA is utilized to produce functional products, primarily proteins.
- Every cell in an organism contains genes, which are segments of DNA that hold the instructions for making specific proteins. Proteins are vital macromolecules that perform a myriad of functions, from providing structural support to cells to catalyzing biochemical reactions. Therefore, the synthesis of the correct proteins in appropriate amounts is crucial for the cell’s survival and proper functioning.
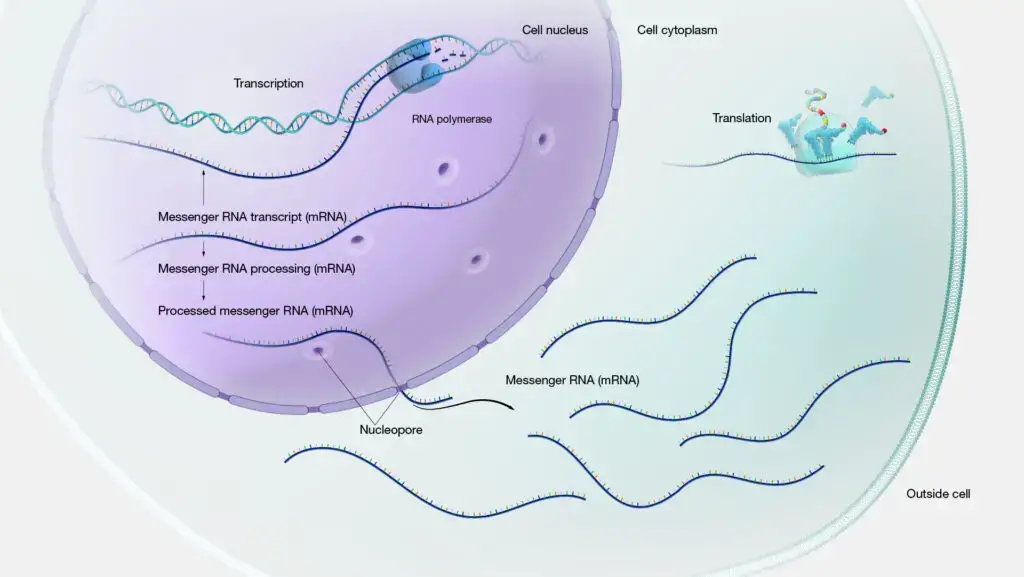
- The process of gene expression begins when a specific gene is activated or “expressed.” This activation results in the production of a molecule called messenger RNA (mRNA). The mRNA serves as a template, carrying the genetic information from the DNA to the cellular machinery responsible for protein synthesis. This step, where the DNA’s information is transcribed into mRNA, is aptly named transcription.
- Following transcription, the mRNA undergoes translation, where its sequence is read and interpreted to assemble amino acids in a specific order, ultimately forming a protein. The sequence in which these amino acids are arranged determines the protein’s structure and, consequently, its function. Besides proteins, some genes produce non-coding RNAs like transfer RNA (tRNA) and small nuclear RNA (snRNA), which play essential roles in various cellular processes.
- It’s imperative to understand that gene expression isn’t a haphazard event. Instead, it’s a highly regulated process, ensuring that proteins are produced when and where they are needed, and in the right quantities. This regulation is pivotal for processes like cellular differentiation, where cells take on specific roles, and morphogenesis, which shapes the form and structure of organisms. Moreover, the adaptability and versatility of an organism hinge on the precise regulation of gene expression. Any deviations or malfunctions in this regulation can lead to diseases and various other complications.
- In summary, gene expression is the bridge between the genetic code and the functional products that determine an organism’s phenotype. It is a meticulously orchestrated process, ensuring that cells produce the necessary proteins to function optimally. Through the intricate dance of transcription and translation, the information stored in genes is brought to life, emphasizing the profound relationship between genotype and phenotype.
Gene expression definition
Gene expression is the process by which the genetic information encoded in a gene is used to produce a functional product, typically a protein or a specific RNA molecule.
Stages in Gene Expression
1. Transcription
- Transcription is a fundamental biological process that serves as the initial phase in the pathway of gene expression. It involves the synthesis of an RNA molecule from a DNA template, effectively converting the genetic information stored in DNA into a format that can be utilized by the cell to produce proteins.
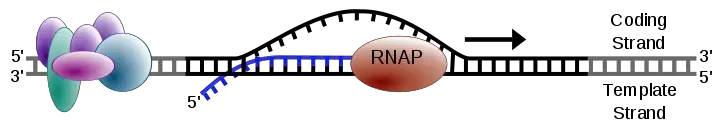
- The process commences when an enzyme known as RNA polymerase binds to a specific region on the DNA, termed the promoter. This promoter region essentially acts as a starting point, signaling the commencement of transcription.
- Once bound, the RNA polymerase initiates the unwinding of the DNA double helix, exposing one of the DNA strands. This exposed strand, referred to as the template strand, guides the synthesis of the RNA molecule.
- As the RNA polymerase progresses along the DNA template, it incorporates RNA nucleotides that are complementary to the DNA sequence. For instance, where the DNA has an adenine (A), the RNA will incorporate a uracil (U). This synthesis proceeds in a 5′ to 3′ direction until the RNA polymerase encounters a termination signal in the DNA.
- Upon reaching this signal, the RNA polymerase detaches from the DNA, and the newly synthesized RNA molecule, known as messenger RNA (mRNA), is released.
- It’s noteworthy to mention that the transcription process varies slightly between prokaryotes and eukaryotes. In prokaryotic cells, transcription and the subsequent step of translation occur simultaneously in the same cellular compartment.
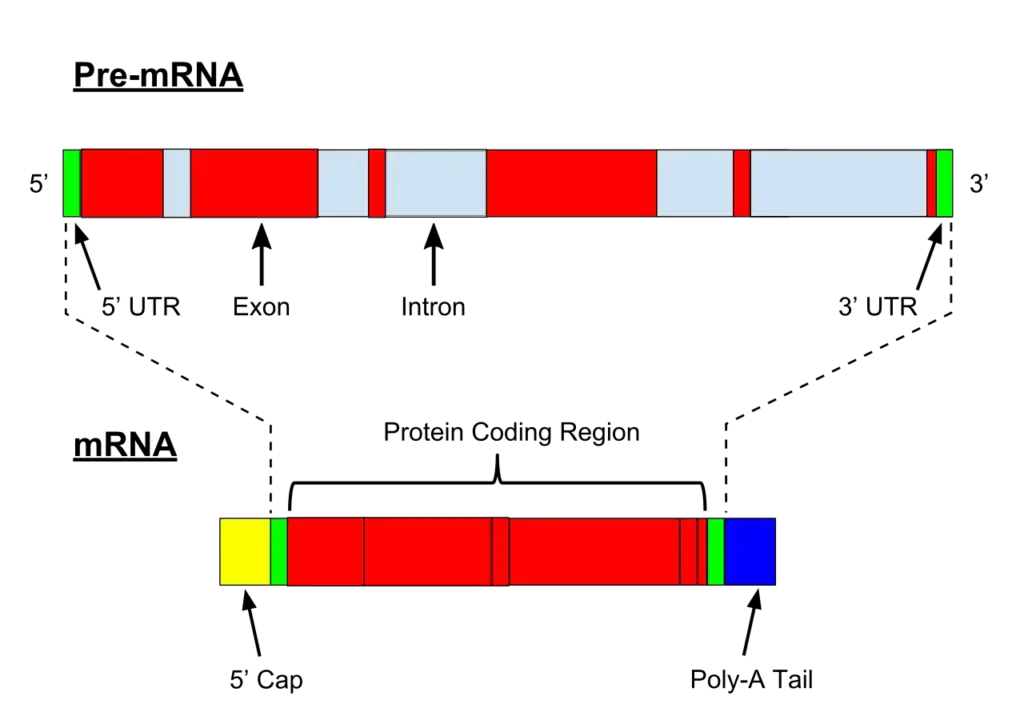
- However, in eukaryotic cells, transcription occurs within the nucleus. The initial RNA transcript, termed pre-mRNA, undergoes various modifications, including capping, splicing, and the addition of a poly-A tail, transforming it into mature mRNA. Following these modifications, the mature mRNA is transported out of the nucleus and into the cytoplasm, where it serves as a template for protein synthesis during the translation process.
- In essence, transcription is the mechanism by which the genetic code within DNA is transcribed into an RNA format, setting the stage for the production of proteins, which are vital for cellular function and structure. Through the meticulous actions of RNA polymerase and associated factors, the information stored in genes is made accessible for the cell’s protein-synthesizing machinery.
2. Translation
- Translation is the cellular process by which the genetic code inscribed in messenger RNA (mRNA) is decoded to produce a specific sequence of amino acids, ultimately forming a protein. This process is a pivotal component of gene expression, ensuring that the information carried by genes is effectively utilized to maintain cellular functionality.
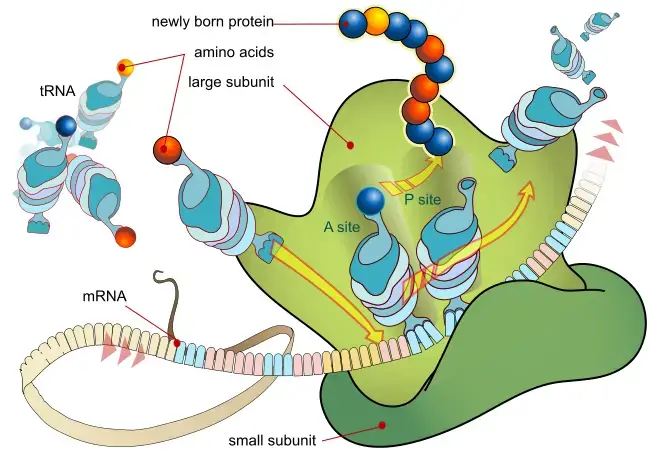
- The translation process commences in the ribosome, a cellular structure that serves as the site for protein synthesis. The ribosome identifies the start codon (AUG) on the mRNA, signaling the beginning of translation. Transfer RNA (tRNA) molecules, each bearing a specific amino acid, play a crucial role in this process. These tRNA molecules possess anticodon sequences that complement the codons on the mRNA. As the ribosome progresses along the mRNA, each codon is paired with its corresponding tRNA anticodon. This ensures that amino acids are assembled in the precise order dictated by the mRNA sequence, forming a growing polypeptide chain.
- The elongation of this polypeptide chain continues until the ribosome encounters a stop codon (UAA, UAG, or UGA) on the mRNA. These stop codons do not correspond to any amino acid; instead, they serve as signals to terminate the translation process. Once the translation is complete, the synthesized protein may undergo various post-translational modifications to achieve its functional form. These modifications can encompass folding into specific structures, addition of functional groups, or segment cleavage, among others.
- It’s essential to note that mRNA molecules can be categorized based on the number of protein sequences they carry. Monocistronic mRNA, commonly found in eukaryotes, codes for a single protein. In contrast, polycistronic mRNA, prevalent in prokaryotes, carries multiple protein sequences.
- Furthermore, the location of translation varies between prokaryotes and eukaryotes. In prokaryotic cells, translation typically occurs concurrently with transcription. In eukaryotic cells, translation can transpire in various regions, such as the cytoplasm or the endoplasmic reticulum, depending on the destined location of the protein.
- In summary, translation is a meticulously coordinated process that deciphers the genetic code in mRNA to produce proteins. Through the synergistic actions of ribosomes, tRNA, and other cellular components, the cell ensures that proteins are synthesized accurately, reflecting the genetic information stored in its DNA.
What is gene expression profiling?
- Gene expression profiling refers to the measurement of the activity or expression levels of genes in a particular cell, tissue, or organism at a given time. It provides valuable insights into how genes are functioning and being regulated under specific conditions, such as during development, disease, or in response to environmental factors.
- Gene expression profiling techniques aim to determine which genes are active (expressed) and to what extent. The expression levels of genes can vary depending on the cell type, tissue type, developmental stage, or disease state. By measuring gene expression, researchers can identify genes that are turned on or off, quantify the amount of gene products (RNA or protein), and compare expression patterns across different samples or conditions.
- There are several methods used for gene expression profiling, including microarrays and next-generation sequencing (NGS) technologies like RNA-Seq. Microarrays involve the immobilization of thousands of known DNA sequences on a solid surface, and the complementary RNA molecules from the sample are labeled and allowed to hybridize with the immobilized DNA probes. This technique provides a snapshot of gene expression by measuring the abundance of specific RNA molecules.
- RNA-Seq, on the other hand, directly sequences the RNA molecules present in a sample. It enables the identification and quantification of all RNA molecules, including known and novel transcripts, providing a more comprehensive view of gene expression. RNA-Seq has become increasingly popular due to its ability to detect rare transcripts, splice variants, and non-coding RNAs.
- The data generated from gene expression profiling experiments can be analyzed using various bioinformatics tools and statistical methods. Researchers can identify differentially expressed genes between conditions, perform clustering analysis to group genes with similar expression patterns, and uncover signaling pathways and biological processes that are activated or repressed.
- Gene expression profiling has a wide range of applications in various fields, including molecular biology, genetics, medicine, and drug discovery. It helps researchers understand the underlying mechanisms of diseases, discover potential biomarkers for diagnosis and prognosis, identify therapeutic targets, and develop personalized medicine approaches.
Methods or Steps of gene expression profiling
Gene expression profiling involves several methods and steps to measure and analyze gene expression levels. Here is a general overview of the process:
- Sample collection: The first step is to collect the biological samples of interest, such as cells, tissues, or organisms. The samples should represent the conditions or experimental factors being investigated.
- RNA extraction: Total RNA is extracted from the collected samples using methods like phenol-chloroform extraction or commercial RNA extraction kits. RNA represents the pool of transcribed genes and serves as the starting material for gene expression profiling.
- RNA quality control: The extracted RNA is checked for quality and integrity using techniques like gel electrophoresis or automated capillary electrophoresis systems. High-quality RNA ensures reliable and accurate downstream analysis.
- cDNA synthesis: Reverse transcription is performed to convert the RNA into complementary DNA (cDNA). This step involves the use of reverse transcriptase enzyme and primers, usually oligo(dT) primers, to generate a pool of cDNA molecules corresponding to the transcribed RNA.
- Labeling of cDNA: If using microarray technology, the cDNA is labeled with fluorescent dyes, such as Cy3 and Cy5, through enzymatic incorporation or chemical coupling. The labeled cDNA samples are then ready for hybridization with microarray chips.
- Microarray hybridization: The labeled cDNA samples are hybridized to the microarray chips containing immobilized DNA probes or oligonucleotides. Each probe on the array corresponds to a specific gene or transcript. The hybridization allows the labeled cDNA to bind to the complementary DNA probes on the array, forming a specific and quantitative measure of gene expression.
- Microarray scanning: After hybridization, the microarray chip is scanned using a specialized scanner that detects the fluorescence signals from the labeled cDNA. The intensities of the fluorescence signals are proportional to the abundance of the corresponding RNA molecules in the original samples.
- Data analysis: The raw data obtained from microarray scanning are subjected to various computational and statistical analyses. These analyses include background subtraction, normalization to correct for technical variations, and statistical tests to identify differentially expressed genes between conditions. Bioinformatics tools and software packages are commonly used for data analysis.
- Validation: To validate the microarray results, selected genes are often further analyzed using independent methods, such as quantitative real-time PCR (qPCR) or RNA sequencing (RNA-Seq). These validation techniques provide more accurate and precise measurements of gene expression levels.
It’s important to note that the advent of next-generation sequencing (NGS) technologies, like RNA-Seq, has expanded gene expression profiling beyond microarrays. RNA-Seq directly sequences the RNA molecules and offers a more comprehensive and unbiased view of gene expression. The steps involved in RNA-Seq differ from microarrays, but they generally include RNA extraction, library preparation, sequencing, and bioinformatic analysis.
Regulation of Gene Expression
Regulation of gene expression is a critical mechanism that ensures the appropriate synthesis of proteins in cells. This regulation is essential for maintaining cellular homeostasis, responding to environmental changes, and guiding developmental processes. By controlling gene expression, cells can produce specific proteins when and where they are needed, ensuring optimal functionality.
- Transcriptional Regulation: The first level of gene expression regulation occurs at the transcription stage. Here, specific proteins called transcription factors bind to DNA sequences, either promoting or inhibiting the binding of RNA polymerase, and consequently, the transcription of genes. Besides these direct genetic interactions, epigenetic modifications play a pivotal role. These modifications, which include DNA methylation and histone alterations, influence the accessibility of genes without altering the DNA sequence itself. Therefore, they can turn genes “on” or “off” based on the cell’s needs.
- Post-transcriptional Regulation: After transcription, the resulting mRNA undergoes various modifications. In eukaryotic cells, segments of the mRNA called introns, which do not code for proteins, are removed. The remaining segments, known as exons, are then joined together. This process, termed splicing, ensures that only the necessary information for protein synthesis remains in the mRNA.
- Translational Regulation: The translation process, where mRNA is used as a template to synthesize proteins, can also be regulated. MicroRNAs, small RNA molecules, can bind to specific sequences in the mRNA, either blocking the initiation of translation or leading to the degradation of the mRNA. Additionally, other molecules, such as translational repressors and RNA-binding proteins, can influence translation by interacting with the mRNA or the translation machinery.
- Post-translational Regulation: Once a protein has been synthesized, it may undergo further modifications to become fully functional. These post-translational modifications can include the addition or removal of specific chemical groups, such as phosphates or acetyl groups. These modifications can alter the protein’s activity, stability, or localization, thereby influencing its role in the cell.
Methods for measuring gene expression
Understanding the levels at which genes are expressed is crucial for gaining insights into various biological processes and phenomena. Several methods have been developed to measure gene expression, each with its own advantages and limitations.
- Northern Blotting: This is one of the traditional methods used to detect specific RNA molecules within a sample. In Northern blotting, RNA is separated using gel electrophoresis and then transferred onto a nitrocellulose membrane. This membrane is then probed with labeled sequences complementary to the target RNA. The strength of the signal provides a measure of the amount of the specific RNA in the sample. While Northern blotting is straightforward and cost-effective, it is also time-consuming and not suitable for high-throughput analyses.
- Quantitative Polymerase Chain Reaction (qPCR): qPCR is a widely used method for quantifying gene expression. It involves the conversion of mRNA to complementary DNA (cDNA) followed by its amplification using PCR. The amplification process is monitored in real-time using fluorescent labels, allowing for the quantification of the starting material. qPCR is known for its sensitivity and speed, but it requires knowledge of the target sequence and can analyze a limited number of targets at once.
- Microarray: This high-throughput method involves the hybridization of cDNA to specific probes attached to a chip. Each spot on the chip corresponds to a specific gene, and the intensity of the signal at each spot indicates the level of expression of that gene. Microarrays can analyze the expression of thousands of genes simultaneously. However, they require specialized equipment for data acquisition and analysis and can sometimes suffer from cross-hybridization issues.
- RNA-Seq: A more recent and advanced method, RNA-Seq involves directly sequencing the RNA from a sample. By counting the number of reads that map to a particular gene, one can infer the gene’s expression level. RNA-Seq offers a broader dynamic range than microarrays and can detect various RNA types, including novel transcripts. However, it is more expensive and demands significant computational resources for data analysis.
What is differential gene expression?
Differential gene expression refers to the phenomenon where genes are expressed at different levels or in different patterns among various cell types, tissues, developmental stages, or in response to different conditions or stimuli. It is a fundamental process that underlies cellular diversity and specialization in multicellular organisms.
Differential gene expression allows cells to acquire distinct identities and perform specific functions within an organism. For example, different cell types in the body, such as neurons, liver cells, and muscle cells, exhibit unique gene expression profiles that enable them to carry out their specialized roles. Similarly, during development, specific genes are activated or repressed in a temporal and spatial manner, guiding the formation of different tissues and organs.
The regulation of differential gene expression is a complex process involving multiple layers of control. It can occur at various stages of gene expression, including transcription, RNA processing, translation, and post-translational modifications. Key regulatory mechanisms involved in generating differential gene expression patterns include:
- Transcriptional regulation: This involves the control of gene expression at the level of transcription. Transcription factors, DNA-binding proteins, and other regulatory elements interact with specific DNA sequences (promoters, enhancers, and silencers) to modulate the binding of RNA polymerase and regulate gene transcription. Differential expression of transcription factors or the presence of specific regulatory elements can lead to different gene expression patterns in different cell types or under different conditions.
- Epigenetic regulation: Epigenetic modifications, such as DNA methylation and histone modifications, can influence gene expression patterns by altering the accessibility of DNA to the transcriptional machinery. These modifications can be heritable and can contribute to the establishment and maintenance of cell type-specific gene expression profiles.
- Post-transcriptional regulation: Differential gene expression can also be achieved through post-transcriptional mechanisms. RNA processing, including alternative splicing and RNA editing, can generate multiple mRNA isoforms from a single gene, each with potentially distinct functions. Additionally, the stability of mRNA molecules can be regulated, affecting their abundance and availability for translation.
- Translational regulation: Once mRNA is produced, its translation into proteins can be regulated. Different cells or conditions may exhibit variations in the efficiency or rate of translation, resulting in differences in protein abundance among cell types.
The study of differential gene expression is essential for understanding the molecular basis of cell differentiation, tissue development, and the response of cells to external cues. It is often investigated using high-throughput techniques such as microarray analysis or RNA sequencing (RNA-seq), which allow for the simultaneous measurement of gene expression levels across thousands of genes. By identifying genes that are differentially expressed, researchers can gain insights into the underlying mechanisms and functional implications of cellular diversity and the regulation of biological processes.
What do You mean by gene expression omnibus?
- The Gene Expression Omnibus (GEO) is a public repository that stores and provides access to gene expression data. It is maintained by the National Center for Biotechnology Information (NCBI), which is part of the United States National Institutes of Health (NIH). GEO serves as a valuable resource for researchers in the field of genomics and provides a platform for sharing and mining gene expression data.
- GEO contains a vast collection of high-throughput gene expression data, including microarray and RNA sequencing (RNA-seq) datasets. These datasets come from a wide range of organisms, including humans, animals, plants, and microorganisms. The data stored in GEO encompasses diverse biological conditions, experimental designs, and research areas, allowing researchers to explore gene expression patterns across different tissues, diseases, developmental stages, and experimental treatments.
- Researchers can deposit their gene expression data in GEO, making it publicly available to the scientific community. This promotes data sharing and enables others to validate and analyze the data, facilitating collaboration and advancing scientific knowledge. GEO provides tools and resources for data retrieval, browsing, and analysis, allowing users to search for specific datasets, perform comparisons between experiments, visualize gene expression patterns, and conduct integrative analyses.
- By accessing GEO, researchers can gain insights into gene expression profiles associated with various biological phenomena, such as disease states, drug treatments, genetic modifications, or environmental factors. The availability of such comprehensive gene expression data sets contributes to a deeper understanding of gene regulation, cellular processes, and the molecular basis of diseases.
- Overall, the Gene Expression Omnibus serves as a vital resource for the scientific community, supporting data sharing, reproducibility, and the discovery of new biological insights through the analysis of gene expression data.
What is taqman gene expression assay?
- The TaqMan gene expression assay is a widely used molecular biology technique for measuring gene expression levels. It is based on the principle of real-time polymerase chain reaction (PCR) and utilizes fluorescent probes to detect and quantify specific RNA transcripts in a sample.
- The assay gets its name from the TaqMan probe, a dual-labeled oligonucleotide probe that is designed to bind to the target RNA sequence of interest. The probe consists of a fluorescent dye (reporter dye) attached to the 5′ end and a quencher molecule attached to the 3′ end. When the probe is intact, the fluorescence of the reporter dye is quenched by the proximity of the quencher molecule.
- During the PCR process, the TaqMan probe is incorporated into the growing DNA strand by the DNA polymerase enzyme. As the DNA strand is amplified, the polymerase reaches the probe binding site. If the target RNA transcript is present in the sample, the probe binds to it and is cleaved by the 5′ to 3′ exonuclease activity of the DNA polymerase. This cleavage separates the reporter dye from the quencher molecule, resulting in an increase in fluorescence intensity.
- The fluorescence emitted by the reporter dye is measured in real time during each cycle of PCR amplification using a specialized instrument called a real-time PCR machine. The fluorescence data is collected and plotted as a fluorescence curve, and the cycle at which the fluorescence signal crosses a predetermined threshold (known as the cycle threshold or Ct value) is used to quantify the initial amount of target RNA in the sample.
- The TaqMan gene expression assay is a highly sensitive and specific method for quantifying gene expression levels. It allows researchers to measure the expression of specific genes in different samples, compare gene expression between experimental groups, and study the dynamics of gene expression under various conditions.
Method of taqman gene expression assay
The TaqMan gene expression assay is a widely used method for quantifying gene expression levels. Here is a step-by-step overview of the assay:
- Designing TaqMan Probes: Specific TaqMan probes are designed to target the RNA sequence of interest. These probes are typically around 20 nucleotides long and are labeled with a fluorescent reporter dye at the 5′ end and a quencher molecule at the 3′ end.
- RNA Extraction: Total RNA is extracted from the cells or tissues of interest using standard RNA extraction methods. This step isolates the RNA molecules, including the target RNA transcript.
- cDNA Synthesis: The extracted RNA is reverse transcribed into complementary DNA (cDNA) using reverse transcriptase enzyme and specific primers. These primers, known as reverse transcription primers, are designed to anneal to the target RNA sequence and initiate the synthesis of cDNA.
- Real-time PCR Setup: The real-time PCR reaction is prepared, which includes the cDNA, gene-specific primers, and the TaqMan probe. The primers are designed to amplify the cDNA corresponding to the target gene, and the TaqMan probe is designed to bind specifically to the target RNA sequence.
- Real-time PCR Amplification: The PCR reaction is subjected to thermal cycling, which includes cycles of denaturation, annealing, and extension. The thermal cycling is performed in a real-time PCR machine that can monitor fluorescence in real time.
- Fluorescence Detection: As the PCR reaction progresses, the TaqMan probe binds to the target RNA sequence if present. During the extension step, the DNA polymerase cleaves the TaqMan probe, separating the reporter dye from the quencher molecule. This cleavage leads to an increase in fluorescence intensity.
- Quantification: The fluorescence emitted by the reporter dye is measured in real time during each cycle of PCR amplification. The cycle threshold (Ct) value, which represents the cycle at which the fluorescence signal crosses a predetermined threshold, is determined. The Ct value is inversely proportional to the amount of target RNA present in the sample.
- Data Analysis: The Ct values obtained from the TaqMan assay can be used to calculate the relative gene expression levels. This is typically done by normalizing the Ct values of the target gene to a reference gene or internal control gene.
The TaqMan gene expression assay provides quantitative information about the expression levels of specific genes in a sample. It is widely used in research and diagnostic applications to study gene expression patterns, identify differentially expressed genes, and validate gene expression changes in various biological systems.
Control of gene expression in prokaryotes
In prokaryotes, the control of gene expression is primarily achieved through the regulation of transcription, which is the process of synthesizing RNA from DNA. Prokaryotes have relatively simple mechanisms for controlling gene expression compared to eukaryotes. Here are some key mechanisms involved in the control of gene expression in prokaryotes:
- Promoter Recognition: Gene expression begins with the binding of RNA polymerase to the promoter region of the DNA. The promoter sequence contains specific DNA elements that allow RNA polymerase to recognize and bind to the correct site for transcription initiation.
- Transcription Factors: Transcription factors are proteins that bind to specific DNA sequences and influence the activity of RNA polymerase. They can either enhance (activators) or inhibit (repressors) transcription. Transcription factors can control gene expression by interacting with RNA polymerase or by affecting the accessibility of the promoter region.
- Operator Region and Operons: In prokaryotes, multiple genes with related functions are often organized together into operons. An operon consists of a promoter region and multiple genes that are transcribed together as a single mRNA molecule. The operator region, located near the promoter, contains binding sites for repressor proteins. The binding of repressor proteins to the operator region can block RNA polymerase from initiating transcription, effectively repressing gene expression.
- Inducible and Repressible Systems: Prokaryotes employ different regulatory systems to respond to changes in the environment. Inducible systems are activated in response to specific signals or inducers. When an inducer molecule binds to a repressor protein, it undergoes a conformational change, releasing the repressor from the operator region, allowing RNA polymerase to initiate transcription. Repressible systems, on the other hand, are continuously active unless repressed by specific corepressor molecules.
- Feedback Inhibition: In some cases, the end product of a metabolic pathway can act as a feedback inhibitor, regulating its own synthesis. The end product binds to a regulatory protein, often an allosteric enzyme, which in turn inhibits the expression of genes involved in its synthesis. This mechanism helps to maintain homeostasis by controlling the production of metabolites based on the cell’s needs.
Overall, the control of gene expression in prokaryotes involves a combination of transcriptional regulation through promoter recognition, transcription factors, and operons, as well as the influence of regulatory proteins, inducers, and feedback inhibition. These mechanisms allow prokaryotes to respond to changes in their environment and regulate the expression of genes necessary for survival and adaptation.
How does dna methylation affect gene expression?
DNA methylation is an epigenetic modification that involves the addition of a methyl group to the DNA molecule, typically at cytosine residues within CpG dinucleotide sequences. DNA methylation can have significant effects on gene expression by influencing the accessibility of genes to the cellular machinery responsible for transcription.
Here’s how DNA methylation affects gene expression:
- Gene Silencing: DNA methylation at the promoter region of a gene can lead to gene silencing or transcriptional repression. When methyl groups are added to the promoter region, they can inhibit the binding of transcription factors and other regulatory proteins to the DNA. This prevents the initiation of transcription, resulting in decreased gene expression. Methylation of promoter regions is often associated with the suppression of gene activity.
- Chromatin Structure: DNA methylation can also influence the structure of chromatin, the complex of DNA and proteins that make up chromosomes. Methyl groups can recruit proteins known as methyl-binding domain (MBD) proteins, which have the ability to modify the structure of chromatin and create a condensed, repressive chromatin state. This compacted chromatin structure makes it difficult for the transcriptional machinery to access the DNA, leading to reduced gene expression.
- Transcription Factor Binding: DNA methylation can directly interfere with the binding of transcription factors to their target sites. Transcription factors are proteins that bind to specific DNA sequences and regulate gene expression. When methylation occurs at the sites where transcription factors are supposed to bind, it can prevent their interaction with the DNA, thereby inhibiting transcriptional activation.
- DNA Stability: DNA methylation can also influence the stability of the DNA molecule itself. Methylated cytosines are more prone to chemical modifications and can undergo spontaneous deamination, leading to the conversion of methylcytosine to thymine. This can result in permanent changes in the DNA sequence and affect gene expression by introducing mutations or disrupting normal DNA structure.
Overall, DNA methylation plays a crucial role in gene expression regulation by modulating the accessibility of genes to the transcriptional machinery. The addition of methyl groups to DNA can lead to gene silencing, chromatin compaction, interference with transcription factor binding, and DNA instability, all of which contribute to the regulation of gene expression patterns in various biological processes and cell types.
Why is gene expression important?
Gene expression is a fundamental process that plays a crucial role in the functioning and development of organisms. Here are several reasons why gene expression is important:
- Protein Synthesis: Gene expression is responsible for the synthesis of proteins, which are the workhorses of cells and perform various functions essential for life. Proteins are involved in nearly all biological processes, including metabolism, cell signaling, structural support, enzyme catalysis, immune response, and many others. Gene expression ensures that the necessary proteins are produced in the right amounts and at the right times to maintain the proper functioning of cells and organisms.
- Cell Differentiation and Development: Gene expression is instrumental in regulating cell differentiation and development. During embryonic development, gene expression determines the specialization of cells into different types, such as nerve cells, muscle cells, or blood cells. The precise regulation of gene expression patterns is crucial for the formation of tissues and organs and the overall development of an organism.
- Response to Environmental Stimuli: Gene expression allows organisms to respond and adapt to changes in their environment. Environmental cues can trigger specific genes to be turned on or off, leading to the production of proteins that help the organism cope with the changing conditions. This ability to regulate gene expression in response to external signals is essential for survival and enables organisms to adjust their metabolism, growth, immune response, and other processes as needed.
- Disease and Health: Aberrant gene expression can lead to various diseases. Dysregulation of gene expression, either through mutations, epigenetic changes, or other factors, can result in the malfunctioning or overproduction of proteins, leading to genetic disorders, developmental abnormalities, or increased susceptibility to diseases such as cancer. Understanding gene expression patterns and mechanisms is crucial for deciphering the underlying causes of diseases and developing effective diagnostic tools and therapeutic interventions.
- Evolution and Diversity: Gene expression is a key driver of evolution and the diversity of life. Changes in gene expression patterns can lead to phenotypic variations, which can be subject to natural selection and contribute to the adaptation of species to their environments. By regulating gene expression, organisms can modify their traits and adapt to different ecological niches, leading to the evolution of new species and the generation of biodiversity.
In summary, gene expression is vital for protein synthesis, cell differentiation, development, environmental responsiveness, disease processes, and the evolution of life. Understanding the mechanisms that control gene expression helps us unravel the complexities of biology, improve our understanding of diseases, and develop new strategies for diagnosis, treatment, and genetic engineering.
Quiz
FAQ
the operon model of the regulation of gene expression in bacteria was proposed by _.
The operon model of the regulation of gene expression in bacteria was proposed by François Jacob and Jacques Monod in 1961. This groundbreaking model was based on their studies of the lac operon in Escherichia coli (E. coli), which demonstrated how genes involved in lactose metabolism are coordinately regulated. Jacob and Monod’s work laid the foundation for understanding gene regulation and provided insights into the principles of transcriptional control in prokaryotes.
protein-phosphorylating enzymes’ role in the regulation of gene expression involves _.
Protein-phosphorylating enzymes play a crucial role in the regulation of gene expression by modulating the activity of transcription factors. Transcription factors are proteins that bind to specific DNA sequences and control the initiation of transcription, the process by which RNA is synthesized from a DNA template. Phosphorylation, the addition of a phosphate group to a protein, can either activate or inactivate transcription factors, thereby influencing gene expression.
The role of protein-phosphorylating enzymes, such as protein kinases, is to transfer phosphate groups from ATP molecules to specific amino acid residues on the transcription factors. This phosphorylation can lead to conformational changes in the transcription factors, affecting their ability to bind to DNA and regulate gene transcription.
Phosphorylation can activate transcription factors by promoting their association with target genes, recruitment of co-activators, or facilitating interactions with other components of the transcriptional machinery. On the other hand, phosphorylation can also result in the inhibition or degradation of transcription factors, preventing their binding to DNA and suppressing gene expression.
Overall, protein-phosphorylating enzymes provide a mechanism for cells to rapidly and reversibly modulate gene expression in response to various signals, including extracellular signals, cellular stress, and developmental cues. By regulating the activity of transcription factors through phosphorylation, these enzymes contribute to the precise control of gene expression in a dynamic and responsive manner.
Epigenetics examines gene expression and how those with identical genes may have different?
Epigenetics is the field of study that examines changes in gene expression and cellular phenotype that are not caused by alterations in the DNA sequence itself. It investigates how identical or similar genetic information can lead to different outcomes or phenotypes in individuals. Epigenetic modifications involve heritable changes in gene expression patterns that do not involve changes in the underlying DNA sequence.
Epigenetic mechanisms play a crucial role in regulating gene expression by influencing the accessibility of genes to the cellular machinery responsible for transcription. They can be influenced by various factors such as environmental cues, lifestyle choices, and developmental processes. Some common epigenetic modifications include DNA methylation, histone modifications, and non-coding RNA molecules.
DNA methylation is a process where a methyl group is added to the DNA molecule, typically to a cytosine residue in a CpG dinucleotide context. DNA methylation can lead to the silencing of gene expression by preventing the binding of transcription factors and other regulatory proteins to the DNA.
Histone modifications involve chemical changes to the proteins called histones, around which DNA is wrapped, and can influence the compactness of the DNA-histone complex. These modifications can either promote or inhibit gene expression by altering the accessibility of the DNA to transcriptional machinery.
Non-coding RNAs, such as microRNAs and long non-coding RNAs, can also regulate gene expression by binding to target mRNA molecules and either promoting their degradation or inhibiting their translation into proteins.
Epigenetic modifications can be heritable, meaning they can be passed on from one generation to another, and they can also be reversible, allowing for dynamic regulation of gene expression throughout an individual’s lifetime.
The study of epigenetics has shed light on how individuals with identical or similar genetic backgrounds can exhibit different phenotypes, such as different susceptibilities to diseases or variations in traits. Epigenetic modifications provide an additional layer of complexity in understanding gene expression regulation and contribute to the diversity and plasticity observed in biological systems.
References
- Volgin, Denys V. (2014). Animal Biotechnology || Gene Expression. , (), 307–325. doi:10.1016/B978-0-12-416002-6.00017-1
- Parker, J. (2001). Brenner’s Encyclopedia of Genetics || Gene Expression. , (), 187–189. doi:10.1016/b978-0-12-374984-0.00587-8
- https://www.genome.gov/genetics-glossary/Gene-Expression#:~:text=Geneexpressionistheprocess,moleculesthatserveotherfunctions.
- https://www.nature.com/scitable/topicpage/gene-expression-14121669/
- https://www.ncbi.nlm.nih.gov/probe/docs/applexpression/
- https://www.cancer.gov/publications/dictionaries/cancer-terms/def/gene-expression
- https://medlineplus.gov/genetics/understanding/howgeneswork/makingprotein/
- https://www.yourgenome.org/facts/what-is-gene-expression/