Flagellar movement, or locomotion helps in the movement of motile bacteria. Bacteria do not drift aimlessly, they move either towards nutrients (sugars and amino acids) or away from toxic substances such as bacterial waste products.
When a bacterial cell moves towards a chemical substance or away from repellents is called chemotaxis. Motile bacteria additionally can shift in response to environmental signals like as oxygen (aerotaxis), osmotic pressure (osmotaxis), temperature (thermotaxis), light (phototaxis), and gravity.
Flagellar Movement
- Bacterial cells move when the filament of bacterial flagella rotates like a propeller, which is trigger by the flagellar motor.
- In E.coli the flagellar motor rotates in 270 rps while in Vibrio alginolyticus averages 1,100 rps.
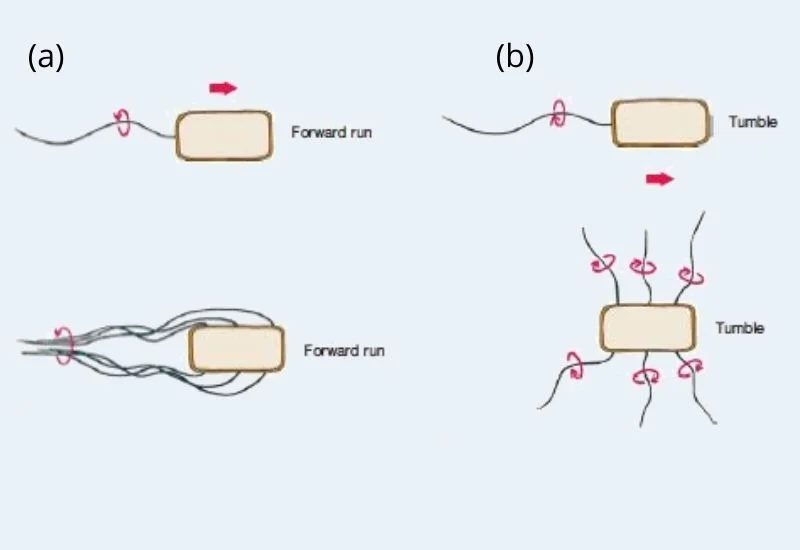
- In aquatic environment, flagellar rotation shows two types of movement such as run and tumble. In run, bacterial cell shows a smooth swimming movement and tumble helps to reorient the cell.
- Bacteria those have monotrichous, polar flagella rotate their filament counterclockwise to moves towards, when the filaments rotate in reverse the cell started to tumbles.
- In Peritrichously flagellated bacteria, the flagella wind at their hooks to create a rotating bundle that drives the cell forward. Clockwise rotation of the flagella interrupts the bundle and the cell tumbles.
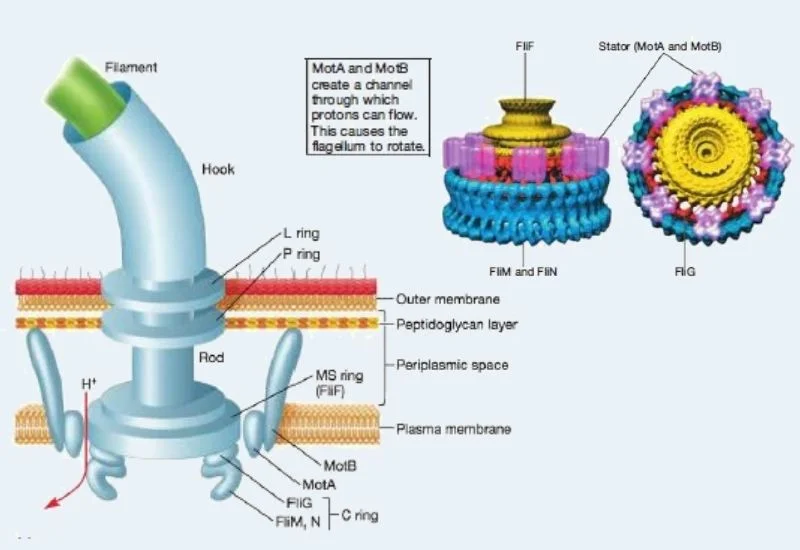
- The rotation of flagella is controlled by the motor which is located at the base of the flagellum. It generates the torque and transmits it to the hook and filament.
- There are two main components of a motor such as the rotor and the stator. It is believed to perform like an electrical motor, where the rotor rotates in the core of a ring of electromagnets, the stator.
- The rotor of gram-negative bacteria is made of MS ring and the C ring. The C ring protein, FliG, is especially significant because it is deemed to combine with the stator.
- The stators are made of two proteins such as MotA and MotB. It helps in the formation of channels through the plasma membrane. MotB also ties MotA to cell wall peptidoglycan.
- The motor uses the proton motive force (PMF) to generates torque and cause flagellar rotation. This power source has a difference in charge and pH across the plasma membrane.
- The proton motive force is mainly generated by the metabolic activities of organisms such as the transfer of electrons from donor to recipient via electron transport chain (ETC).
- In ETC protons transported to the outside of the cell from the cytoplasm as a result the outside has more positively charged ions (the protons) and a lower pH.
- PMF is a type of possible energy that can be utilized to do work such as mechanical work, as in the case of flagellar rotation; transport work, the movement of substances inside or out of the cell; or chemical work like as the synthesis of ATP, the cell’s energy currency.
Mechanism of Flagellar Movement
The channels produced by the MotA and MotB proteins permit protons to pass beyond the plasma membrane from the surface to the interior.
Therefore, the protons flow down the charge and pH gradient. This action frees energy that is utilized to rotate the flagellum. In reality, the entrance of a proton within the channel is alike the entrance of a person into a spinning door.
The “power” of the proton creates torque, somewhat same as a person driving the spinning door. Certainly, the rate of flagellar rotation is proportional to the quantity of the PMF.
Types of Bacterial Motility
There are five types of movement which are been observed within bacterial cell such as;
- Swimming movement conferred by flagella
- Flagella-mediated swarming
- Corkscrew movement of spirochetes.
- Twitching motility associated with type IV pili.
- Gliding motility.
Swimming
- In aquatic environment, bacteria show swimming movement by using flagella.
- Swimming can be reversible or unidirectional.
- Swimming is a challenging task for bacteria due to the viscosity of the surrounding water seems.
- The cell needs to penetrate through the water by its corkscrew-shaped flagella, and if flagellar movement stops, it freezes almost immediately.
- In this type of environment, bacteria can move from 20 to approximately 90 µm per second.
- Example: Swimming motility is found in Pseudomonas aeruginosa.
Swarming
- A rising amount of bacterial species has been observed to manifest an exceptional type of motility known as swarming.
- The Swarming motility only found on moist surfaces and is a variety of group performance in which cells migrate in communities across the surface.
- Most of the bacteria those have Swarming motility contain peritrichous flagella. Many of them also create and secrete molecules that assist them to travel across the substrate.
- Example: This motility has been studied in genus Serratia, Salmonella, Aeromonas, Bacillus, Yersinia, Pseudomonas, Proteus, Vibrio and Escherichia.
Spirochete Motility
- Spirochete exhibits a distinct type of motility. They contain multiple flagella that arise from different ends of the cell.
- These flagella are intertwined and wind around the cell.
- These flagella are not extended to the outside of the cell wall but preferably settle in the periplasmic space and are covered by the outer membrane.
- They are known as periplasmic flagella and are believed to revolve same as the external flagella of other bacteria, producing the corkscrew-shaped outer membrane to revolve and depart the cell within the surrounding liquid, even very viscous waters.
- When spirochetes come in touch with a solid surface Flagellar rotation started to flex or bend the cell and as a result, it shows the creeping or crawling movement.
Twitching
- Twitching motility is defined by little, irregular, jerky movements of up to several micrometers in length and is usually observed on very wet surfaces.
- Bacteria show Twitching motility on solid surface.
- In Twitching motility, Involve type IV pili, the production of slime, or both. The type IV pili are found at one or both poles of bacteria.
- The pili are estimated to alternately stretch and retreat to depart bacteria through twitching motility.
- The elongated pilus reaches the surface at a point any way from the cell body. When the pilus retreats, the cell is dragged ahead.
- This extension/retraction process is powered by the hydrolysis of ATP.
- Example: Twitching motility is found in Pseudomonas aeruginosa, Neisseria gonorrhoeae and Myxococcus xanthus
Gliding Motility
- Gliding motility is soft and alters greatly in rate (from 2 to over 600 flm per minute) and in the nature of the motion.
- Bacteria either glide along in a direction parallel to the longitudinal axis of their cells or move with a screwlike motion or even migrate in a direction perpendicular to the long axis of the cells.
- During gliding, others rotate around their longitudinal axis.
- Example: The gliding motility id found in Flavobacterium spp. and Mycoplasma spp.
Chemotaxis
- In Chemotaxis nutrients acts as attractant near the bacterial population the bacteria will go towards it and on the other hand when there is any toxic substance it acts as a repellent. In its vicinity, the bacterial population will go away from it so this is how the chemotaxis works in bacteria
- But how this attractant and repellent are censored within the bacteria? will be shown in the following signaling pathway.
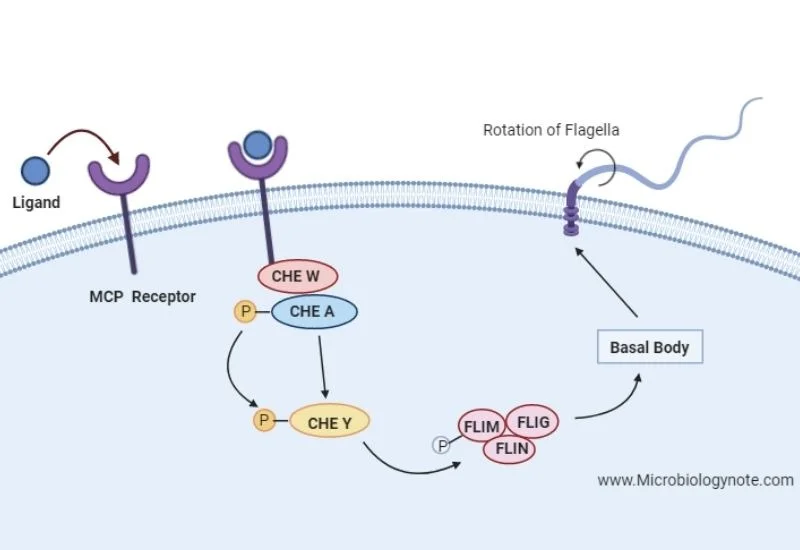
- The chemotaxis pathway is media to through a transmembrane protein called MCP methyl accepting chemotaxis protein. It’s a receptor protein which is able to detect with the repellents and attractants in the environment prior to ligand binding.
- The receptor is in the form of monomers and when there is any substance a repellent near the bacteria the repellent acts as a ligand for the MCP receptor. The repellent acts as a ligand for MCP receptor and shows ligand binding activity towards this receptor.
- The binding of ligand towards receptor is followed by the dimerization of MCP. So now the mCP has got conformational changes in its structure that means it has got activated by a ligand. So after dimerization of MCP the MCP recruits two more proteins one is called CHE W or CW protein and another one is CHE A protein or you can say CA protein.
- The CW protein only acts as it turns to sir protein for the signal to be transmitted to CA protein and it is this CA protein which acts as a sensor kinase. The CA shows kinase activity and Auto phosphorylates its hysteria residues.
- After the auto phosphorylation of CA protein the CA protein activates the CHE Y protein by transferring the phosphate towards the aspartate residue off CHE Y. Finally the CHE Y protein postulates the protein complex composed of three proteins FLIM, FLIG, and FLIN.
- This protein complex acts as a pledge learn more to switch and when this CHE Y protein activates this switch it gives signal to basal body of the flagella to switch the rotation for direction change.
- In this case the ligand was repellent so the rotation of flagella will be switching from counterclockwise to clockwise rotation to get away from the substance that’s toxic for bacteria.
- Now let’s see the movements in bacteria depending upon the substances which bacteria encounter. First of all we see bacteria showing forward movements in presence of nutrients and at that time the flagellum rotation is counterclockwise that means the bacteria is going forward.
- But when there is the presence of any toxic chemicals in its vicinity the counter-clockwise movement will switch it to the clockwise rotation of the flagellum and the bacteria will go away from that substance. This is all about chemotaxis in bacteria.