What is FASTA format?
FASTA format is a text-based format commonly used for representing nucleotide or protein sequences. It is named after the FASTA software package, which introduced this format. The FASTA format is widely used in bioinformatics and genomics for storing and exchanging sequence data. FASTA file looks like:

A typical FASTA file consists of one or more entries, each representing a sequence along with its associated information. Here is the general structure of a FASTA entry:
- A single-line description or identifier starting with a greater-than symbol (“>”), followed by optional additional information about the sequence.
- One or more lines containing the sequence data.
For example, a FASTA entry representing a DNA sequence might look like this:
sequence_identifier
ATCGATCGATCGATCGATCGATCGATCGIn this example, the line starting with “>” represents the sequence identifier, which can be a unique identifier or a descriptive label for the sequence. The subsequent line contains the actual sequence data. The sequence can be written in a single line or divided into multiple lines for readability, although each line is typically limited to a certain character length.
FASTA format can also be used for protein sequences, in which case the sequence is represented by the standard one-letter amino acid codes. For example:
>protein_identifier
MNIFEMLRIDEGLRLKIYKDTEGYYTIGIGHLLTKSPSLNAAKSELDKAIGRNTNGVITKDEAEKLFNQDVDAAVRGILRNAKLKPVYDSLDAVRRAALEAHPELARDA
FASTA files can contain multiple entries, with each entry starting with a “>” line followed by the sequence data. This allows the format to store and organize multiple sequences within a single file.
The simplicity and widespread usage of FASTA format make it easy to work with and share sequence data among different bioinformatics tools and databases.
Principle of FASTA Formate
FASTA is a pairwise sequence alignment tool that compares input sequences of nucleotides or proteins with existing databases. This format is text-based and can be read and written using a text editor or word processor. On a single line, a ‘>’ symbol precedes the gi and accession number, which are followed by the description of a Fasta file. Each row would only contain 60 nucleotides/amino acids. The sequence would begin on the next line, and each row would consist of 60 nucleotides/amino acids. For DNA and proteins, IUPAC nucleotide codes and amino acid codes are represented by a single letter. It determines the sequences’ local similarity and calculates the statistical significance of matches. It can also be used to determine the evolutionary and functional relationship between sequences.
Before attempting a more time-consuming optimized search, the FASTA program employs word hits to identify potential matches. Speed and sensitivity are determined by the parameter ktup, which specifies the length of each word. By increasing the ktup, the quantity of background hits is reduced. Initially, it looks for segments with multiple adjacent hits. This program is significantly more sensitive than BLAST programs, as evidenced by the duration of time needed to generate results. FASTA generates local alignment scores for the comparison of the query sequence to all database sequences. This method avoids the artificiality of a random sequence model by employing actual sequences with their inherent correlations. The sequences are obtained using the subsequent procedures.
DNA sequencing methods
- Sanger Method (dideoxy chain termination method): The Sanger technique (dideoxy chain termination technique) involves labeling four test tubes with the letters A, T, G, and C. Each test instrument requires the addition of denatured DNA (single strands). Next, a primer that anneals to one of the strands in the template is added. The 3′ end of the primer contains both the dideoxy nucleotides (unique to each tube) and the deoxy nucleotides arbitrarily. When ddNTPs are added to a growing chain, the chain ends due to a paucity of 3’OH, which forms a phosphodiester bond with the next nucleotide. Thus, tiny DNA strands are formed. The sequence order can be determined by analyzing the patterns in the gel based on their molecular weight following electrophoresis. Additionally, the primer or one of the nucleotides can be radioactively or fluorescently labeled, allowing the final product to be readily detected on the gel and the sequence to be inferred.
- Maxam-Gilbert (Chemical degradation method): This method also requires denatured DNA and the radioactive modification of the 5′ end of the DNA strand, as well as the purification of the DNA fragment. Through chemical treatment, a series of labeled fragments are generated. After electrophoresis, fragments are organized in a gel. To observe the fragments, the gel is exposed to X-ray film for autoradiography, and a series of dark bands correspond to radiolabeled DNA fragments from which the sequence can be inferred.
Protein sequencing methods
- Edman Degradation reaction: The reaction determines the order of amino acids in a protein from the N-terminus by cleaving each amino acid from the N-terminus without disrupting the protein’s bonds. After each cleavage, the amino acid is identified by chromatography or electrophoresis.
- Mass Spectrometry: It is used to determine the mass of particles, the composition of molecules, and the chemical structures of molecules, such as peptides and other chemical compounds. On the basis of the mass-to-charge ratio, the amino acids in a protein can be identified.
FASTA Programs
The FASTA programs provide a range of functionalities for sequence alignment, database searching, motif discovery, and other sequence-related tasks. Here are some popular FASTA programs:
- FASTA: The original FASTA program developed by William Pearson. It performs pairwise sequence alignments using heuristics to efficiently search large sequence databases.
- FASTX and FASTY: These programs are similar to FASTA but specifically designed for comparing nucleotide sequences (FASTX) or translated nucleotide sequences against protein databases (FASTY).
- TFASTX and TFASTY: These programs are accelerated versions of FASTX and FASTY, respectively, optimized for comparing DNA sequences to protein databases.
- BLAST (Basic Local Alignment Search Tool): Although not explicitly named after the FASTA format, BLAST is a widely used sequence similarity search tool that shares similarities with the FASTA programs. It includes various flavors such as BLASTN (nucleotide-nucleotide), BLASTP (protein-protein), BLASTX (translated nucleotide-protein), and more.
- PSI-BLAST: Position-Specific Iterated BLAST is an enhanced version of BLAST that performs iterative searches to identify distantly related protein sequences.
- HMMER: Although not based on the FASTA format, HMMER is a popular tool for analyzing sequences using Hidden Markov Models (HMMs). It provides functionalities for profile hidden Markov model (HMM) searches, building custom HMMs, and more.
- EMBOSS: The European Molecular Biology Open Software Suite (EMBOSS) is a comprehensive collection of bioinformatics tools that includes various programs for sequence analysis. Some EMBOSS tools, such as “needle” and “water,” perform pairwise sequence alignments similar to FASTA.
- SSEARCH: SSEARCH (Smith-Waterman SEARCH) is a program developed by William Pearson that performs local sequence alignments using the Smith-Waterman algorithm. It is similar to the original FASTA program but uses a more rigorous algorithm to identify local similarities between sequences.
- GGSEARCH/GLSEARCH: GGSEARCH and GLSEARCH are programs within the FASTA package that perform global sequence alignments. GGSEARCH is optimized for comparing protein sequences, while GLSEARCH is designed for nucleotide sequences. They use the Needleman-Wunsch algorithm to find the optimal global alignment between two sequences.
- FASTS/ TFASTS: FASTS and TFASTS are accelerated versions of the original FASTA program and TFASTX, respectively. They are optimized for searching sequence databases using short, ungapped motifs or patterns. FASTS is used for protein queries, while TFASTS is designed for translated nucleotide sequences.
- FASTF/ TFASTF: FASTF and TFASTF are another pair of programs within the FASTA package. They are similar to FASTS and TFASTS but are optimized for searching sequence databases using short, gapped motifs or patterns. FASTF is used for protein queries, and TFASTF is designed for translated nucleotide sequences.
These programs are widely used in bioinformatics research and have contributed significantly to sequence analysis, alignment, and database searching tasks. Each program has its own unique features and strengths, allowing researchers to choose the most appropriate tool for their specific analysis needs.
Parameters used in FASTA algorithm
- Threshold: The threshold parameter in the FASTA algorithm determines the minimum score or similarity value required for a sequence alignment to be considered significant. Alignments below this threshold are usually filtered out as noise or random matches. The specific threshold value depends on the scoring system and significance level chosen by the user.
- True Homology: True homology refers to the genuine evolutionary relationship between two sequences. In the context of FASTA, the goal is to identify true homologous sequences that share a common ancestor. By comparing sequences and applying statistical analysis, FASTA attempts to identify alignments that are likely to represent true homology rather than chance matches.
- Putative Conserved Domains: Conserved domains are functional and structurally important regions within a protein sequence that are evolutionarily conserved across related sequences. FASTA can be used to identify putative conserved domains by detecting sequence similarities that span these regions. The presence of such conserved domains can provide insights into the functional and evolutionary relationships of proteins.
- Scoring Matrix: The scoring matrix defines the values assigned to matches and mismatches between residues in the sequence alignment. The most commonly used scoring matrix is the BLOSUM matrix series for protein sequences, such as BLOSUM62 or BLOSUM50. For nucleotide sequences, a simple match/mismatch scoring is often used.
- Gap Penalties: Gap penalties determine the cost of introducing gaps (insertions or deletions) in the alignment. They include parameters for opening a gap (gap opening penalty) and extending an existing gap (gap extension penalty). The values of these parameters affect the alignment’s sensitivity to insertions and deletions.
- Word Size: FASTA employs a heuristic approach based on word matching to identify potential sequence similarities. The word size parameter specifies the length of the word (substring) used for matching. A larger word size increases sensitivity but also requires more computational resources.
- Expectation Value (E-value): The E-value is a statistical parameter that estimates the number of sequence alignments expected to occur by chance in a database search. A lower E-value threshold indicates a more significant similarity. Users can set the E-value threshold to control the level of significance required for reporting matches.
- Filter Options: FASTA provides various filtering options to improve the efficiency of the search process. For example, the low complexity filter identifies and masks regions of low complexity before alignment. Other filters, such as the repeat filter, remove redundant or repetitive sequences from the database.
- Output Format: The FASTA algorithm allows users to specify the desired output format for the sequence alignments and search results. This includes options for displaying the alignment scores, sequence identifiers, alignment positions, and other relevant information.
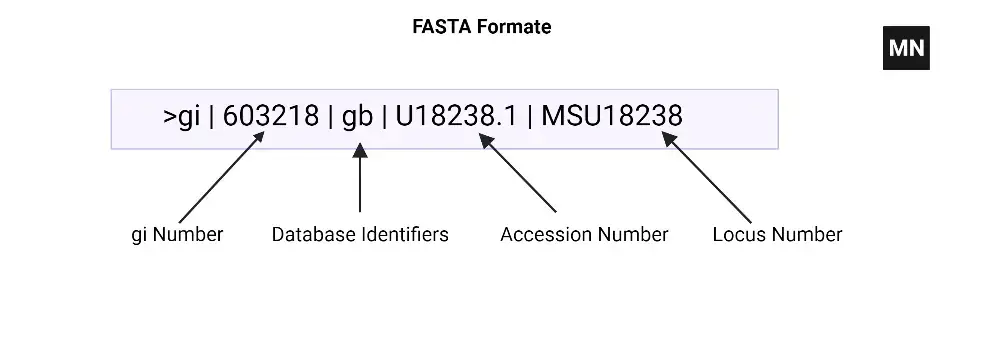
FASTA Formate
The FASTA format consists of two main parts: the sequence identifier line (header) and the sequence data.

- Sequence Identifier Line (Header):
- The sequence identifier line starts with a “>” (greater-than) symbol, followed by a unique identifier or description of the sequence.
- The identifier can be a simple alphanumeric string or a more detailed description, such as the name of the organism, gene, or protein.
- Additional information can be included after the identifier, such as database accession numbers or annotations, separated by spaces or other delimiters.
- Example: >KJ946236.1 Homo sapiens Kidd blood group protein (SLC14A1) gene, exons 4, 5 and partial cds
- Sequence Data:
- The sequence data follows the sequence identifier line and consists of the actual sequence of nucleotides (for DNA or RNA) or amino acids (for proteins).
- The sequence can span multiple lines and should only contain the valid characters corresponding to the type of sequence (e.g., A, T, G, C for DNA).
- The sequence can be represented in either uppercase or lowercase letters, although uppercase is more commonly used.
- There should be no line breaks or spaces within the sequence data.
- Example: ATGGAGGACAGCCCCACTATGGTTAGAGTGGACAGCCCCACTATGGTTAGGGGTGAAAACCAGGTTTCGCCATGTCAAGGGAGAAGGTGCTTCCCCAAAGCTCTTGGCTATGTCACCGGTGACATGAAAAAACTTGCCAACCAGCTTAAAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNACAAACCCGTGGTGCTCCAGTTCATTGACTGGATTCTCCGGGGCATATCCCAAGTGGTGTTCGTCAACGACCCCGTCAGTGGAATCCTGATTCTGGTAGGACTTCTTGTTCAGAACCCCTGGTGGGCTCTCACTGGCTGGCTGGGAACAGTGGTCTCCACTCTGATGGCCCTCTTGCTCAGCCAGGACAG
The FASTA format allows multiple sequences to be included in a single file, where each sequence is represented by a sequence identifier line followed by its corresponding sequence data. Multiple sequences can be used for various purposes, such as database searches, multiple sequence alignments, or batch processing.
It’s important to note that while the basic structure of the FASTA format remains consistent, there can be variations in how specific software or databases implement additional features or annotations within the sequence identifier line.
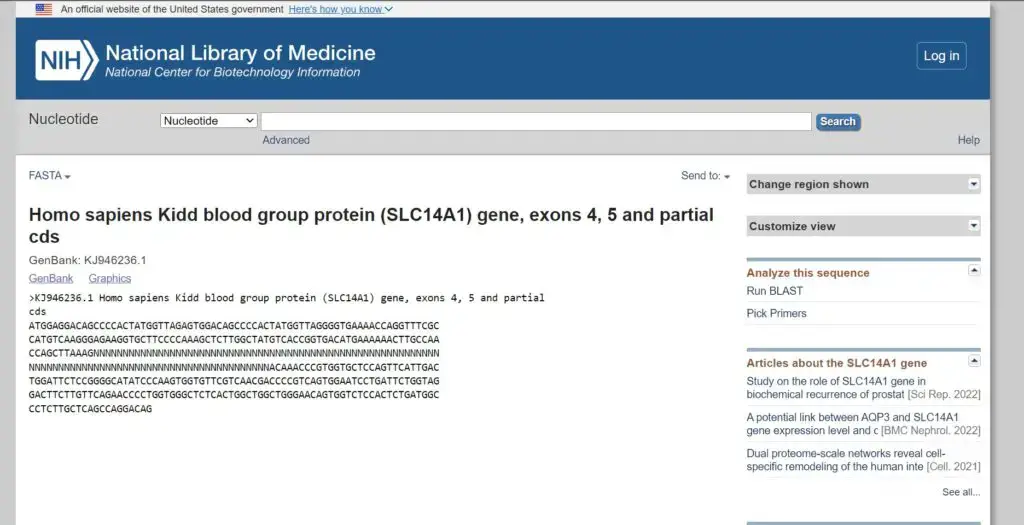
How to Get FASTA Formate From NCBI?
To obtain FASTA format sequences from the NCBI (National Center for Biotechnology Information) database, you can follow these steps:
- Go to the NCBI website (https://www.ncbi.nlm.nih.gov) and access the desired database, such as the NCBI Nucleotide (NT) database or the NCBI Protein database.
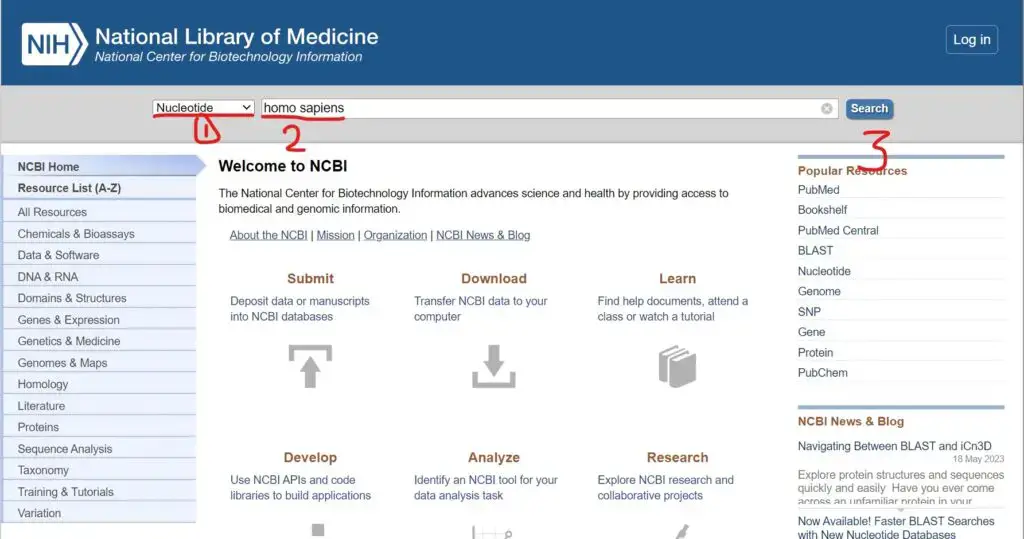
- Perform a search for the sequence(s) of interest using keywords, accession numbers, gene names, or any other relevant identifiers.
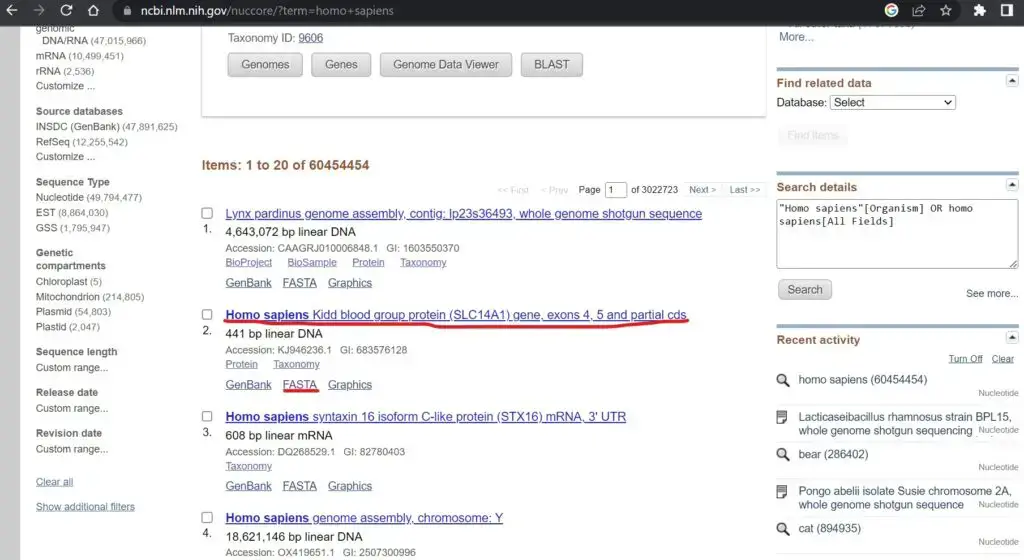
- From the search results, select the specific sequence(s) you want to download in FASTA format. You can use checkboxes or other selection options provided by NCBI to choose multiple sequences if needed.
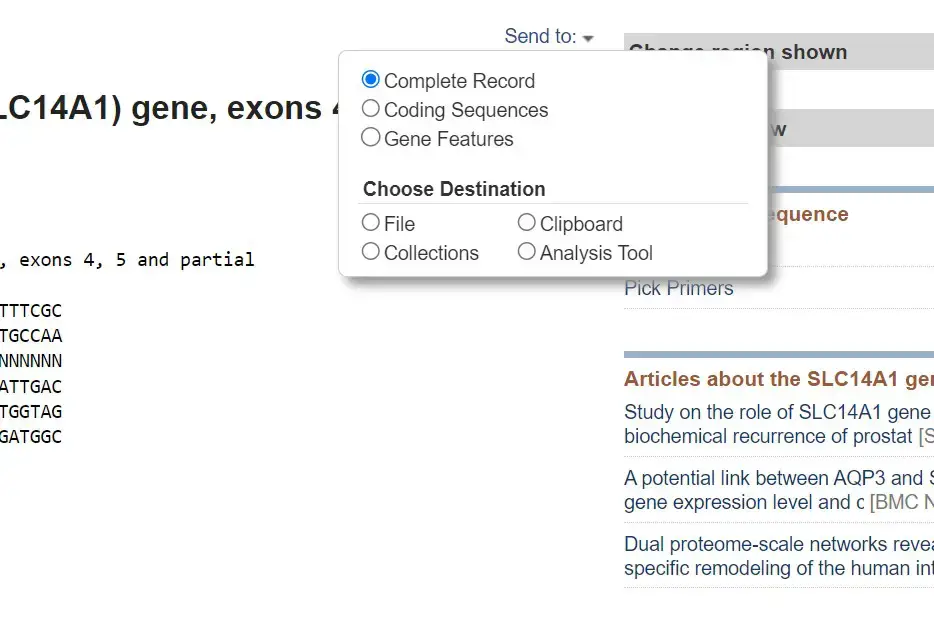
- Once you have selected the desired sequences, look for a button or link that allows you to download the sequences. In most cases, there will be a “Send to” or “Download” option.
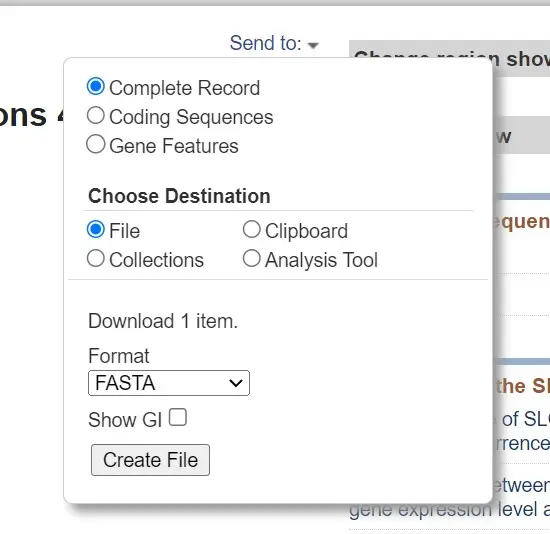
- Click on the appropriate option to access the download menu. From there, choose the “FASTA” format as the desired format for download. You may also have options to select specific database records or customize the download.
- After selecting the FASTA format, click the “Download” or “Submit” button to initiate the download process.
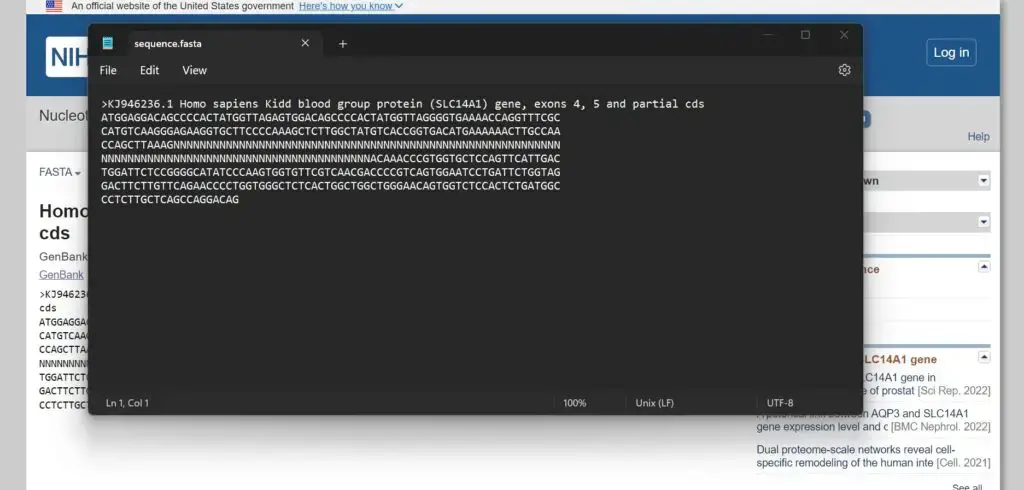
- The sequences will be downloaded as a text file in FASTA format. You can open the file using a text editor to view the sequences in the desired FASTA format.
Procedure to run FASTA program
The procedure for executing a FASTA program may vary marginally based on the specific implementation or application being used. However, the general procedures are as follows:
- Install and Configure: Ensure that the FASTA software or program is installed on your computer. It may be necessary to obtain and install it from the appropriate source using the provided installation instructions.
- Prepare Input Files: Prepare the required input files for FASTA analysis. Typically, a query sequence file and a target sequence database file are included. The query sequence file contains the search sequence for the target database. The target database file contains the sequences you wish to compare the query against.
- Define Parameters: Identify the parameters desired for the FASTA analysis. This includes specifying the scoring matrix, penalty for gaps, word length, and any other pertinent parameters. Consider the characteristics of your sequences and the objectives of your analysis when establishing these parameters.
- Run the Program: Open the FASTA program’s command-line interface or graphical user interface (GUI). Provide the required command or select the appropriate GUI options to initiate an analysis. Specify the required input files and parameter values.
- Monitor Progress: During the execution of the FASTA program, you may see indicators of progress or status updates. The execution duration can vary depending on the size of the database and the complexity of the analysis. Monitor the program’s progress to ensure its seamless operation.
- Analyze Results: Upon completion of the FASTA analysis, the program will generate output files comprising the results. Typically, these files contain sequence alignments, alignment scores, statistical measures (such as the E-value), and other pertinent data. Retrieve the output files and examine them to analyze and interpret the results.
- Post-processing and Interpretation: Depending on the specific objectives of the analysis, it may be necessary to perform additional post-processing or downstream analysis on the FASTA results. This may involve the visualization, filtering, statistical analysis, or integration with other tools or databases in order to obtain additional insights into the sequence similarities and their biological significance.
How FASTA Works
To identify and align sequence similarities, the FASTA algorithm employs a heuristic approach. Here is an overview of FASTA’s operation:
- Preprocessing: Prior to beginning the search, the target sequence database is preprocessed to generate an index for effective searching. This indexing step entails dividing the database sequences into fixed-length fragments or words (commonly referred to as “seeds” or “words”). These words and their corresponding positions in the original sequences are preserved in a data structure, such as a hash table or suffix tree.
- Query Sequence Comparison: Using a word-based matching strategy, the query sequence is compared against the sequences of the target database. Additionally, the query sequence is divided into words of the same length as those used in the indexing process.
- Word Matching: The FASTA algorithm begins by identifying precise matches between the query sequence’s words and the target database’s words. It searches through the query sequence for terms that match any of the database’s words. When a match is discovered, an alignment extension is initiated.
- Alignment Extension: After identifying a word match, FASTA extends the alignment in both directions, iteratively checking for matches and calculating a score based on the selected scoring matrix and gap penalties. The extension of the alignment continues until the score falls below a predetermined threshold or no further extensions are conceivable.
- Scoring and Scoring Matrix: During alignment extension, FASTA assigns scores to residue matches, discrepancies, and gap introductions in accordance with the selected scoring matrix (e.g., BLOSUM for proteins). These scores contribute to the aggregate alignment score, which reflects the sequence similarity between the query and the target.
- Statistical Significance: After calculating an alignment score, FASTA estimates the statistical significance of the alignment by calculating an Expectation Value (E-value). In a random sequence, the E-value represents the expected number of alignments with the same or higher score that would occur by chance.
- Output and Post-processing: FASTA reports alignments whose scores surpass a specified significance threshold (E-value). Depending on the implementation or program used, the output typically includes the alignment itself, the alignment score, statistical measures, and other pertinent information.
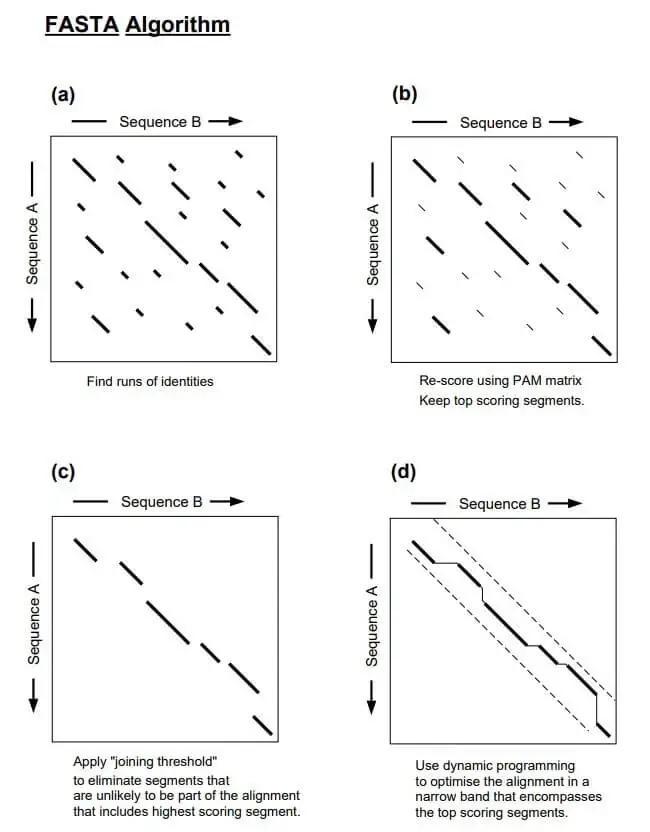
It is essential to note that the precise details and variations of the FASTA algorithm may vary based on the specific implementation or application. However, the fundamental concept of word matching, alignment extension, scoring, and estimation of statistical significance remains consistent across the majority of FASTA implementations.
Statistical Significance and FASTA
In bioinformatics, statistical significance is an essential concept, including in the context of the FASTA algorithm. When conducting sequence database searches with FASTA, statistical significance helps assess the likelihood that a sequence alignment or similarity observed between a query sequence and a database sequence is due to chance as opposed to a meaningful biological similarity.
The FASTA algorithm calculates a score for each alignment it discovers, which reflects the similarity between the sequences of the query and the database. This result is determined by the scoring matrix and gap penalties selected. FASTA calculates an Expectation Value (E-value) to ascertain the statistical significance of the alignment. The E-value approximates the number of alignments that might occur by coincidence during a database search.
A lower E-value indicates a more substantial alignment, indicating a greater likelihood of true sequence homology. Typically, researchers use an E-value threshold to eliminate alignments that are likely random or spurious.
Noting that the E-value is a statistical estimate and not an absolute measure of significance is essential. It is the expected number of alignments with the same or a higher score that could occur by coincidence. It is therefore essential to interpret the E-value within the context of the specific analysis and the nature of the sequences being compared.
In conclusion, statistical significance, as assessed by the E-value in the FASTA algorithm, assists researchers in determining the dependability and biological significance of sequence alignments. It enables the identification of meaningful similarities between sequences and helps to differentiate them from coincidental database matches.
Advantages of FASTA
- Sensitivity: FASTA is known for its sensitivity in detecting sequence similarities, including subtle relationships between sequences that may be missed by other methods. It employs heuristics and statistical models that allow for efficient and accurate identification of homologous sequences.
- Flexibility: The FASTA algorithm offers a wide range of options and parameters, allowing users to tailor the analysis to their specific needs. It supports different scoring matrices, gap penalties, and filtering options, providing flexibility in sequence alignment and database searching.
- Speed: FASTA is designed to handle large sequence databases efficiently. It employs indexing techniques and heuristic algorithms that expedite the search process, making it well-suited for analyzing large-scale genomic and proteomic datasets.
- Versatility: FASTA programs cover a broad spectrum of sequence analysis tasks, including pairwise alignment, motif discovery, database searching, and more. This versatility makes it a valuable tool for various applications in bioinformatics and computational biology.
- Established and Widely Used: FASTA has been widely used in the bioinformatics community for several decades. It has a well-established reputation, a large user base, and extensive support from the research community. This makes it easier to find resources, documentation, and assistance when working with FASTA.
Limitations of FASTA
- Computational Resources: The sensitivity and flexibility of FASTA come at the cost of increased computational requirements. Searching large databases or performing exhaustive comparisons can be computationally intensive and may require substantial computing resources.
- Execution Time: While FASTA is generally faster than the more rigorous Smith-Waterman algorithm, it may still be slower compared to some newer algorithms. As sequence databases continue to grow, the execution time for searching the entire database may become a limiting factor.
- Statistical Significance Estimation: The E-value calculated by FASTA provides an estimate of statistical significance, but it is based on assumptions and approximations. The interpretation of E-values requires caution, as they are subject to various statistical considerations and can vary depending on the specific parameters chosen.
- Sequence Length Limitation: Some versions of FASTA may have limitations on the maximum sequence length that can be processed. This can be a constraint when dealing with extremely long sequences, such as whole-genome sequences.
- Dependency on Database Quality: The quality and completeness of the sequence databases used for comparison impact the performance of FASTA. Incomplete or outdated databases may affect the accuracy and reliability of the results.
Applications of FASTA
The FASTA algorithm and associated programs have numerous bioinformatics and sequence analysis applications. Here are some common FASTA applications:
- Sequence Database Searching: FASTA is commonly used for searching sequence databases in order to identify similarities between a query sequence and sequences in a database. It facilitates the identification of homologous sequences, the detection of evolutionary relationships, and the inference of functional annotations.
- Sequence Alignment: FASTA performs both local and global pairwise sequence alignments, enabling researchers to compare and align two sequences. This is beneficial for identifying conserved regions, discovering functional domains, and researching sequence variations.
- Motif Discovery: FASTA can be utilized to discover conserved motifs or patterns within sequences. Researchers can identify putative functional elements in protein or nucleotide sequences by looking for short, ungapped motifs (using FASTS/TFASTS) or gapped motifs (using FASTF/TFASTF).
- Homology Modeling: FASTA can assist in homology modeling, which predicts the structure of an unknown protein or nucleotide sequence by using a known homologous structure as a template.
- Comparative Genomics: FASTA is indispensable for comparative genomics research. It facilitates the comparison of genomic sequences across species, the identification of orthologous genes, and the study of evolutionary relationships between organisms.
- Functional Annotation: FASTA facilitates the functional annotation of genes and proteins by identifying sequence similarities and homologous relationships. It assists in the assignment of putative functions to newly sequenced genes based on similarities to previously characterized sequences.
- Metagenomics: FASTA is used to analyze complex microbial communities in metagenomic investigations by comparing sequenced reads or contigs to reference databases. It facilitates the identification and characterization of microbial taxa present in a sample.
- Analysis of Next-Generation Sequencing (NGS) Data: FASTA programs are frequently employed in the preprocessing and analysis of NGS data. They assist with quality control, read mapping, variant calling, and other duties associated with the analysis of large sequencing datasets.
- Homolog Identification: Identification of Homologous Sequences: FASTA can be used to identify homologous sequences across multiple organisms or species. Researchers can identify related sequences that share a common ancestor by comparing a query sequence against a database of sequences. This information is essential for comprehending evolutionary relationships and researching gene families.
- Phylogenetic Analysis: FASTA can contribute to phylogenetic analysis by supplying sequence alignments for use in constructing phylogenetic trees. By aligning homologous sequences and inferring evolutionary relationships, FASTA facilitates the comprehension of the evolution of various organisms and their genetic relatedness.
- Primer Design: FASTA can aid in the design of primers for polymerase chain reaction (PCR) experiments. By aligning the sequence of a primer against a database of target sequences, researchers can evaluate the primer’s specificity and potential for amplification. This facilitates precise and specific PCR amplification.
- Functional Conservation Analysis: FASTA can be used to analyze the conservation of functional domains or motifs across multiple species. Researchers can obtain insight into the functional significance and evolutionary conservation of specific protein or nucleotide motifs by aligning sequences and identifying conserved regions.
- Comparative Transcriptomics: In transcriptomic investigations, FASTA can facilitate the comparison of transcript sequences across various samples or conditions. Researchers can identify differentially expressed genes, discover alternative splicing events, and investigate gene expression patterns by aligning and comparing RNA sequences.
- Protein Structure Prediction: Prediction of Protein Structure FASTA can be used to predict protein structure by identifying homologous sequences with known structures. By aligning the target protein sequence with homologous proteins of known structures, researchers can infer the target protein’s likely structure and generate structural models.
- Drug Discovery: FASTA contributes to the drug discovery process by facilitating the identification of potential drug targets. Researchers can identify prospective candidate proteins for drug development by comparing the protein sequences of known drug targets with databases of potential target proteins.
Things to Remerber when use FASTA
- Sequence Quality: Ensure that the input sequences, both the query sequence and the sequences in the database, are error-free and of high quality. Low-quality sequences can lead to erroneous or misleading conclusions.
- Parameter Selection: Understanding the impact of various FASTA algorithm parameters, such as the scoring matrix, gap penalties, and filtering options, is required for parameter selection. Choose appropriate parameter values based on the characteristics of your sequences and your analysis objectives. It may be necessary to experiment with various parameter settings in order to achieve optimal results.
- Statistical Significance: Estimates of statistical significance, such as the E-value, should be interpreted with caution. In spite of the fact that they provide a measure of significance, they are founded on assumptions and approximations. Consider the E-value threshold with consideration and take the context and nature of the analysis into account.
- Database Selection: For your analysis, select a relevant and up-to-date sequence database. The accuracy and dependability of the results can be substantially impacted by the database’s quality and comprehensiveness. Be conscious of any potential biases or restrictions associated with the selected database.
- Benchmarking: If possible, validate the FASTA results using independent methodologies or benchmark datasets. The precision and dependability of the FASTA output can be evaluated by comparing the results to known or experimentally validated similarities.
- Sensitivity of Parameters: Be mindful that the selected parameters, specifically the word size and interval penalties, can influence the sensitivity of FASTA. Adjusting these parameters can affect the algorithm’s ability to detect sequence similarities and its overall performance.
- Resource Considerations: FASTA searches can be computationally intensive, depending on the extent of the sequence database and the computational complexity of the analysis. Ensure that you have sufficient computational resources, such as processing capacity, memory, and storage, to efficiently handle the analysis.
- Result Interpretation: When interpreting the results, you should consider the biological context and the specific objectives of your analysis. Evaluation of sequence similarity should take into account the known biology, functional annotations, and evolutionary relationships. Consider using additional tools and resources to analyze and validate the results further.
FAQ
What is FASTA and what is its purpose in bioinformatics?
FASTA is a widely used algorithm and suite of programs in bioinformatics. Its purpose is to search for sequence similarities, perform sequence alignments, and facilitate various sequence analysis tasks.
How does the FASTA algorithm work?
The FASTA algorithm utilizes a heuristic approach, combining word matching, alignment extension, scoring, and statistical significance estimation to identify and align sequence similarities.
What are the different programs associated with the FASTA algorithm?
Some of the commonly used programs associated with FASTA include the original FASTA program, SSEARCH, GGSEARCH/GLSEARCH, FASTS/TFASTS, and FASTF/TFASTF.
What are the main applications of FASTA in sequence analysis?
FASTA has diverse applications, including homolog identification, phylogenetic analysis, primer design, functional conservation analysis, comparative transcriptomics, protein structure prediction, and drug discovery.
How do I choose the appropriate scoring matrix and gap penalties for a FASTA analysis?
The choice of scoring matrix and gap penalties depends on the type of sequences being analyzed (protein or nucleotide) and the specific goals of the analysis. Commonly used scoring matrices for proteins include BLOSUM and PAM matrices.
What is the significance of the E-value reported by FASTA?
The E-value provides an estimate of the statistical significance of the sequence alignment. It represents the expected number of alignments with the same or better score that would occur by chance in a random sequence.
Can FASTA handle large-scale sequence databases?
Yes, FASTA is designed to handle large sequence databases efficiently. It utilizes indexing techniques and heuristic algorithms to expedite the search process, making it suitable for analyzing large-scale genomic and proteomic datasets.
How can I interpret the sequence alignment results generated by FASTA?
Sequence alignment results from FASTA can be interpreted by analyzing the alignment score, E-value, sequence identities, and positions of conserved residues. The biological context and specific goals of the analysis should also be considered when interpreting the results.
Are there any limitations or potential pitfalls when using FASTA?
Some limitations of FASTA include the computational resources required, sensitivity to parameter selection, statistical assumptions in estimating significance, potential sequence length limitations, and dependency on the quality and completeness of the sequence databases used.
What are some alternative sequence analysis algorithms or methods to consider alongside FASTA?
Some alternative sequence analysis algorithms to consider alongside FASTA include BLAST (Basic Local Alignment Search Tool), HMMER (Hidden Markov Model-based sequence analysis), ClustalW/MUSCLE for multiple sequence alignment, and various machine learning-based approaches for sequence analysis.
References
- Pearson, W. R. (2013). An Introduction to Sequence Similarity (“Homology”) Searching. Current Protocols in Bioinformatics, 42(1), 3.1.1-3.1.8.
- Pearson, W. R., & Lipman, D. J. (1988). Improved tools for biological sequence comparison. Proceedings of the National Academy of Sciences, 85(8), 2444-2448.
- Lipman, D. J., & Pearson, W. R. (1985). Rapid and sensitive protein similarity searches. Science, 227(4693), 1435-1441.
- Altschul, S. F., Gish, W., Miller, W., Myers, E. W., & Lipman, D. J. (1990). Basic local alignment search tool. Journal of Molecular Biology, 215(3), 403-410.
- Mott, R., & Zvelebil, M. J. (1994). PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research, 22(17), 3389-3402.
- Pearson, W. R., & Miller, W. (1992). Dynamic programming algorithms for biological sequence comparison. Methods in Enzymology, 210, 575-601.
- Thompson, J. D., Higgins, D. G., & Gibson, T. J. (1994). CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research, 22(22), 4673-4680.
- Gertz, E. M., & Yu, Y. K. (2003). Agglomerative clustering of a search engine query log. Proceedings of the 12th International Conference on World Wide Web, 407-416.
- Jones, D. T. (1999). Protein secondary structure prediction based on position-specific scoring matrices. Journal of Molecular Biology, 292(2), 195-202.
- Madeira, F., & Park, Y. M. (2019). Protein similarity networks reveal relationships among sequence, structure, and function. Science Advances, 5(1), eaau9739.
- Chen, Zhen & Zhao, Pei & Li, Fuyi & Marquez-Lago, Tatiana & Leier, Andre & Revote, Jerico & Zhu, Yan & Powell, David & Akutsu, Tatsuya & Webb, Geoffrey & Chou, Kuo-Chen & Smith, A & Daly, Roger & Li, Jian & Song, Jiangning. (2020). iLearn: an integrated platform and meta-learner for feature engineering, machine learning analysis and modeling of DNA, RNA and protein sequence data. Briefings in Bioinformatics. 21. 1047–1057. 10.1093/bib/bbz041.
- Hosseini, Morteza & Pratas, Diogo & Pinho, Armando. (2016). A Survey on Data Compression Methods for Biological Sequences. information, MDPI. 7. 56. 10.3390/info7040056.
- https://www.ncbi.nlm.nih.gov/genbank/fastaformat/#:~:text=In%20FASTA%20format%20the%20line,should%20not%20contain%20any%20spaces.
- Text Highlighting: Select any text in the post content to highlight it
- Text Annotation: Select text and add comments with annotations
- Comment Management: Edit or delete your own comments
- Highlight Management: Remove your own highlights
How to use: Simply select any text in the post content above, and you'll see annotation options. Login here or create an account to get started.